Example Dysmalpy 2D fitting
In this example, we use dysmalpy to measure the kinematics of galaxy GS4_43501 at \(z=1.613\). This notebook shows how to find the best fit models for the two-dimensional velocity and velocity dispersion profiles using \(\texttt{MPFIT}\) as well as \(\texttt{MCMC}\). These fits allow us to measure quantities such as the total mass (disk+bulge), the effective radius \(r_\mathrm{eff}\), dark matter fraction \(f_\mathrm{DM}\) and velocity dispersion \(\sigma_0\).
The fitting includes the following components:
Disk + Bulge
NFW halo
Constant velocity dispersion
The structure of the notebook is the following:
Setup steps
Initialize galaxy, model set, instrument
MPFIT fitting
MCMC fitting
Visualize a real MCMC example
Nested sampling fitting
Visualize a real Nested Sampling example
1) Setup steps
Import modules
from __future__ import (absolute_import, division, print_function,
unicode_literals)
import dysmalpy
from dysmalpy import galaxy
from dysmalpy import models
from dysmalpy import fitting
from dysmalpy import instrument
from dysmalpy import data_classes
from dysmalpy import parameters
from dysmalpy import plotting
from dysmalpy import aperture_classes
from dysmalpy import observation
from dysmalpy import config
import os
import copy
import numpy as np
import astropy.units as u
import astropy.io.fits as fits
INFO:numexpr.utils:Note: NumExpr detected 10 cores but "NUMEXPR_MAX_THREADS" not set, so enforcing safe limit of 8.
INFO:numexpr.utils:NumExpr defaulting to 8 threads.
# A check for compatibility:
import emcee
if int(emcee.__version__[0]) >= 3:
ftype_sampler = 'h5'
else:
ftype_sampler = 'pickle'
Setup notebook
# Setup plotting
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
mpl.rcParams['figure.dpi']= 100
mpl.rc("savefig", dpi=300)
from IPython.core.display import Image
import logging
logger = logging.getLogger('DysmalPy')
logger.setLevel(logging.INFO)
Set data, output paths
# Data directory
dir_path = os.path.abspath(fitting.__file__)
data_dir = os.sep.join(os.path.dirname(dir_path).split(os.sep)[:-1]+["tests", "test_data", ""])
#'/YOUR/DATA/PATH/'
print(data_dir)
# Where to save output files (see examples below)
#outdir = '/Users/sedona/data/dysmalpy_test_examples/JUPYTER_OUTPUT_2D/'
outdir = '/Users/jespejo/Dropbox/Postdoc/Data/dysmalpy_test_examples/JUPYTER_OUTPUT_2D/'
/Users/jespejo/anaconda3/envs/test_dysmalpy/lib/python3.11/site-packages/dysmalpy/tests/test_data/
Load the function to tie the scale height to the disk effective radius
from dysmalpy.fitting_wrappers.tied_functions import tie_sigz_reff
Load the function to tie Mvirial to \(f_{DM}(R_e)\)
from dysmalpy.fitting_wrappers.tied_functions import tie_lmvirial_NFW
Info
Also see fitting_wrappers.tied_functions for more tied functions
2) Initialize galaxy, model set, instrument
gal = galaxy.Galaxy(z=1.613, name='GS4_43501')
mod_set = models.ModelSet()
Baryonic component: Combined Disk+Bulge
total_mass = 11.0 # M_sun
bt = 0.3 # Bulge-Total ratio
r_eff_disk = 5.0 # kpc
n_disk = 1.0
invq_disk = 5.0
r_eff_bulge = 1.0 # kpc
n_bulge = 4.0
invq_bulge = 1.0
noord_flat = True # Switch for applying Noordermeer flattening
# Fix components
bary_fixed = {'total_mass': False,
'r_eff_disk': False, #True,
'n_disk': True,
'r_eff_bulge': True,
'n_bulge': True,
'bt': True}
# Set bounds
bary_bounds = {'total_mass': (10, 13),
'r_eff_disk': (1.0, 30.0),
'n_disk': (1, 8),
'r_eff_bulge': (1, 5),
'n_bulge': (1, 8),
'bt': (0, 1)}
bary = models.DiskBulge(total_mass=total_mass, bt=bt,
r_eff_disk=r_eff_disk, n_disk=n_disk,
invq_disk=invq_disk,
r_eff_bulge=r_eff_bulge, n_bulge=n_bulge,
invq_bulge=invq_bulge,
noord_flat=noord_flat,
name='disk+bulge',
fixed=bary_fixed, bounds=bary_bounds)
bary.r_eff_disk.prior = parameters.BoundedGaussianPrior(center=5.0, stddev=1.0)
Halo component
mvirial = 12.0
conc = 5.0
fdm = 0.5
halo_fixed = {'mvirial': False,
'conc': True,
'fdm': False}
# Mvirial will be tied -- so must set 'fixed=False' for Mvirial...
halo_bounds = {'mvirial': (10, 13),
'conc': (1, 20),
'fdm': (0, 1)}
halo = models.NFW(mvirial=mvirial, conc=conc, fdm=fdm, z=gal.z,
fixed=halo_fixed, bounds=halo_bounds, name='halo')
halo.mvirial.tied = tie_lmvirial_NFW
Dispersion profile
sigma0 = 39. # km/s
disp_fixed = {'sigma0': False}
disp_bounds = {'sigma0': (5, 300)}
disp_prof = models.DispersionConst(sigma0=sigma0, fixed=disp_fixed,
bounds=disp_bounds, name='dispprof', tracer='halpha')
z-height profile
sigmaz = 0.9 # kpc
zheight_fixed = {'sigmaz': False}
zheight_prof = models.ZHeightGauss(sigmaz=sigmaz, name='zheightgaus',
fixed=zheight_fixed)
zheight_prof.sigmaz.tied = tie_sigz_reff
Geometry
inc = 62. # degrees
pa = 142. # degrees, blue-shifted side CCW from north
xshift = 0 # pixels from center
yshift = 0 # pixels from center
vel_shift = 0 # velocity shift at center ; km/s
geom_fixed = {'inc': False,
'pa': False,
'xshift': False,
'yshift': False,
'vel_shift': False}
geom_bounds = {'inc': (52, 72),
'pa': (132, 152),
'xshift': (-2.5, 2.5),
'yshift': (-2.5, 2.5),
'vel_shift': (-100, 100)}
geom = models.Geometry(inc=inc, pa=pa, xshift=xshift, yshift=yshift, vel_shift=vel_shift,
fixed=geom_fixed, bounds=geom_bounds,
name='geom', obs_name='halpha_2D')
geom.inc.prior = parameters.BoundedSineGaussianPrior(center=62, stddev=0.1)
Add all model components to ModelSet
# Add all of the model components to the ModelSet
mod_set.add_component(bary, light=True)
mod_set.add_component(halo)
mod_set.add_component(disp_prof)
mod_set.add_component(zheight_prof)
mod_set.add_component(geom)
Set kinematic options for calculating velocity profile
mod_set.kinematic_options.adiabatic_contract = False
mod_set.kinematic_options.pressure_support = True
Set up the observation and instrument
obs = observation.Observation(name='halpha_2D', tracer='halpha')
inst = instrument.Instrument()
beamsize = 0.55*u.arcsec # FWHM of beam
sig_inst = 45*u.km/u.s # Instrumental spectral resolution
beam = instrument.GaussianBeam(major=beamsize)
lsf = instrument.LSF(sig_inst)
inst.beam = beam
inst.lsf = lsf
inst.pixscale = 0.125*u.arcsec # arcsec/pixel
inst.fov = [27, 27] # (nx, ny) pixels
inst.spec_type = 'velocity' # 'velocity' or 'wavelength'
inst.spec_step = 10*u.km/u.s # Spectral step
inst.spec_start = -1000*u.km/u.s # Starting value of spectrum
inst.nspec = 201 # Number of spectral pixels
# Set the beam kernel so it doesn't have to be calculated every step
inst.set_beam_kernel()
inst.set_lsf_kernel()
# Extraction information
inst.ndim = 2 # Dimensionality of data
inst.moment = False # For 1D/2D data, if True then velocities and dispersion calculated from moments
# Default is False, meaning Gaussian extraction used
# Add instrument to observation
obs.instrument = inst
Load data
Load the data from file:
2D velocity, dispersion maps and error
A mask can be loaded / created as well
Put data in
Data2DclassAdd data to Galaxy object
gal_vel = fits.getdata(data_dir+'GS4_43501_Ha_vm.fits')
gal_disp = fits.getdata(data_dir+'GS4_43501_Ha_dm.fits')
err_vel = fits.getdata(data_dir+'GS4_43501_Ha_vm_err.fits')
err_disp = fits.getdata(data_dir+'GS4_43501_Ha_dm_err.fits')
mask = fits.getdata(data_dir+'GS4_43501_Ha_m.fits')
#gal_disp[(gal_disp > 1000.) | (~np.isfinite(gal_disp))] = -1e6
#mask[(gs4_disp < 0)] = 0
inst_corr = True # Flag for if the measured dispersion has been
# corrected for instrumental resolution
# Mask NaNs:
mask[~np.isfinite(gal_vel)] = 0
gal_vel[~np.isfinite(gal_vel)] = 0.
mask[~np.isfinite(err_vel)] = 0
err_vel[~np.isfinite(err_vel)] = 0.
mask[~np.isfinite(gal_disp)] = 0
gal_disp[~np.isfinite(gal_disp)] = 0.
mask[~np.isfinite(err_disp)] = 0
err_disp[~np.isfinite(err_disp)] = 0.
# Put data in Data2D data class:
# ** specifies data pixscale as well **
data2d = data_classes.Data2D(pixscale=inst.pixscale.value, velocity=gal_vel,
vel_disp=gal_disp, vel_err=err_vel,
vel_disp_err=err_disp, mask=mask,
inst_corr=inst_corr)
# Use moment for extraction for example:
# Not how data was extracted, but it's a speedup for this example.
#
# The best practic option, if the 2D maps are extracted with gaussians,
# is to compile the C++ extensions and pass the option 'gauss_extract_with_c=True'
# when running fitting.fit_mpfit()
data2d.moment = True
# Add data to Observation:
obs.data = copy.deepcopy(data2d)
Define model, fit options:
obs.mod_options.oversample = 1
# Factor by which to oversample model (eg, subpixels)
# For increased precision, oversample=3 is suggested.
obs.fit_options.fit = True # Include this observation in the fit (T/F)
obs.fit_options.fit_velocity = True # 1D/2D: Fit velocity of observation (T/F)
obs.fit_options.fit_dispersion = True # 1D/2D: Fit dispersion of observation (T/F)
obs.fit_options.fit_flux = False # 1D/2D: Fit flux of observation (T/F)
Add the model set, observation to the Galaxy
gal.model = mod_set
gal.add_observation(obs)
3) MPFIT Fitting
Set up MPFITFitter fitter with fitting parameters
# Options passed to MPFIT:
maxiter = 200
fitter = fitting.MPFITFitter(maxiter=maxiter)
Set up fit/plot output options
# Output directory
outdir_mpfit = outdir+'MPFIT/'
# Output options:
do_plotting = True
plot_type = 'png'
overwrite = True
output_options = config.OutputOptions(outdir=outdir_mpfit,
do_plotting=do_plotting,
plot_type=plot_type,
overwrite=overwrite)
Run Dysmalpy fitting: MPFIT
mpfit_results = fitter.fit(gal, output_options)
INFO:DysmalPy:*************************************
INFO:DysmalPy: Fitting: GS4_43501 using MPFIT
INFO:DysmalPy: obs: halpha_2D
INFO:DysmalPy: nSubpixels: 1
INFO:DysmalPy: mvirial_tied: <function tie_lmvirial_NFW at 0x176a55da0>
INFO:DysmalPy:
MPFIT Fitting:
Start: 2024-05-02 10:47:55.313966
INFO:DysmalPy:Iter 1 CHI-SQUARE = 27260.87785 DOF = 403
disk+bulge:total_mass = 11
disk+bulge:r_eff_disk = 5
halo:fdm = 0.5
dispprof:sigma0 = 39
geom:inc = 62
geom:pa = 142
geom:xshift = 0
geom:yshift = 0
geom:vel_shift = 0
INFO:DysmalPy:Iter 2 CHI-SQUARE = 7174.918633 DOF = 403
disk+bulge:total_mass = 11.01621875
disk+bulge:r_eff_disk = 5.697081099
halo:fdm = 0.290454085
dispprof:sigma0 = 41.78914475
geom:inc = 72
geom:pa = 142.4487865
geom:xshift = 0.01555573571
geom:yshift = -0.01483259239
geom:vel_shift = 0.688311908
INFO:DysmalPy:Iter 3 CHI-SQUARE = 4845.547462 DOF = 403
disk+bulge:total_mass = 11.00485748
disk+bulge:r_eff_disk = 7.299527889
halo:fdm = 0.3316062539
dispprof:sigma0 = 35.80898768
geom:inc = 72
geom:pa = 142.6165451
geom:xshift = 0.00498880543
geom:yshift = -0.01907966512
geom:vel_shift = 1.26046722
INFO:DysmalPy:Iter 4 CHI-SQUARE = 4135.278964 DOF = 403
disk+bulge:total_mass = 10.97290392
disk+bulge:r_eff_disk = 5.906465959
halo:fdm = 0.2268716489
dispprof:sigma0 = 39.19735991
geom:inc = 72
geom:pa = 142.9235737
geom:xshift = 0.006769462096
geom:yshift = -0.04087333416
geom:vel_shift = 3.910223933
INFO:DysmalPy:Iter 5 CHI-SQUARE = 2681.1139 DOF = 403
disk+bulge:total_mass = 10.99187836
disk+bulge:r_eff_disk = 5.260076359
halo:fdm = 0.06304476313
dispprof:sigma0 = 35.0014785
geom:inc = 72
geom:pa = 144.7019383
geom:xshift = 0.005638298412
geom:yshift = -0.08458184636
geom:vel_shift = 13.29443606
INFO:DysmalPy:Iter 6 CHI-SQUARE = 1975.039779 DOF = 403
disk+bulge:total_mass = 11.01752643
disk+bulge:r_eff_disk = 5.972062778
halo:fdm = 0.07041719829
dispprof:sigma0 = 33.79762307
geom:inc = 72
geom:pa = 144.82705
geom:xshift = 0.003838933716
geom:yshift = -0.08965685046
geom:vel_shift = 22.53154176
INFO:DysmalPy:Iter 7 CHI-SQUARE = 1911.611202 DOF = 403
disk+bulge:total_mass = 11.02762255
disk+bulge:r_eff_disk = 6.236754678
halo:fdm = 0.07706825794
dispprof:sigma0 = 35.78878353
geom:inc = 72
geom:pa = 145.0908239
geom:xshift = 0.002765540566
geom:yshift = -0.08522907649
geom:vel_shift = 24.69604616
INFO:DysmalPy:Iter 8 CHI-SQUARE = 1874.721763 DOF = 403
disk+bulge:total_mass = 11.05717868
disk+bulge:r_eff_disk = 6.756909482
halo:fdm = 0.08265623498
dispprof:sigma0 = 36.07195236
geom:inc = 72
geom:pa = 145.1249566
geom:xshift = 0.002925563871
geom:yshift = -0.09434522976
geom:vel_shift = 24.74136223
INFO:DysmalPy:Iter 9 CHI-SQUARE = 1868.205783 DOF = 403
disk+bulge:total_mass = 11.07330048
disk+bulge:r_eff_disk = 6.976880571
halo:fdm = 0.08303211692
dispprof:sigma0 = 39.11660957
geom:inc = 72
geom:pa = 145.0231882
geom:xshift = 0.002306094135
geom:yshift = -0.1027435952
geom:vel_shift = 24.51481928
INFO:DysmalPy:Iter 10 CHI-SQUARE = 1834.891502 DOF = 403
disk+bulge:total_mass = 11.12364485
disk+bulge:r_eff_disk = 7.783313281
halo:fdm = 0.05379420088
dispprof:sigma0 = 35.87322102
geom:inc = 72
geom:pa = 145.0823943
geom:xshift = 0.002328402216
geom:yshift = -0.1072879964
geom:vel_shift = 24.3734785
INFO:DysmalPy:Iter 11 CHI-SQUARE = 1804.57498 DOF = 403
disk+bulge:total_mass = 11.15233343
disk+bulge:r_eff_disk = 8.49550137
halo:fdm = 0.05314505098
dispprof:sigma0 = 35.22862485
geom:inc = 72
geom:pa = 145.2209654
geom:xshift = 0.00213117077
geom:yshift = -0.1118302835
geom:vel_shift = 25.7821227
INFO:DysmalPy:Iter 12 CHI-SQUARE = 1765.875956 DOF = 403
disk+bulge:total_mass = 11.1995498
disk+bulge:r_eff_disk = 9.671163543
halo:fdm = 0.0567955085
dispprof:sigma0 = 33.6639676
geom:inc = 72
geom:pa = 145.1054693
geom:xshift = 0.001771156208
geom:yshift = -0.1214819783
geom:vel_shift = 25.15294892
INFO:DysmalPy:Iter 13 CHI-SQUARE = 1750.678931 DOF = 403
disk+bulge:total_mass = 11.23072663
disk+bulge:r_eff_disk = 10.71370417
halo:fdm = 0.06951073943
dispprof:sigma0 = 33.20391113
geom:inc = 72
geom:pa = 145.5238662
geom:xshift = 0.001667999645
geom:yshift = -0.1229693221
geom:vel_shift = 25.23407271
INFO:DysmalPy:Iter 14 CHI-SQUARE = 1749.269119 DOF = 403
disk+bulge:total_mass = 11.20953224
disk+bulge:r_eff_disk = 10.26453885
halo:fdm = 0.07783456653
dispprof:sigma0 = 33.20999322
geom:inc = 72
geom:pa = 145.2975363
geom:xshift = 0.00150466758
geom:yshift = -0.1259508646
geom:vel_shift = 25.64745219
INFO:DysmalPy:Iter 15 CHI-SQUARE = 1742.263826 DOF = 403
disk+bulge:total_mass = 11.26350202
disk+bulge:r_eff_disk = 11.69794866
halo:fdm = 0.06604308184
dispprof:sigma0 = 30.21624009
geom:inc = 72
geom:pa = 145.2839822
geom:xshift = 0.001305084891
geom:yshift = -0.1274298711
geom:vel_shift = 24.36968127
INFO:DysmalPy:Iter 16 CHI-SQUARE = 1739.337454 DOF = 403
disk+bulge:total_mass = 11.27240108
disk+bulge:r_eff_disk = 12.14000166
halo:fdm = 0.06225635304
dispprof:sigma0 = 29.57297226
geom:inc = 72
geom:pa = 145.3862099
geom:xshift = 0.001253914203
geom:yshift = -0.1281740116
geom:vel_shift = 25.17378889
INFO:DysmalPy:Iter 17 CHI-SQUARE = 1739.259112 DOF = 403
disk+bulge:total_mass = 11.27160988
disk+bulge:r_eff_disk = 12.16727606
halo:fdm = 0.06791349056
dispprof:sigma0 = 29.6275998
geom:inc = 72
geom:pa = 145.5798563
geom:xshift = 0.00125420715
geom:yshift = -0.1281740091
geom:vel_shift = 25.16057188
INFO:DysmalPy:Iter 18 CHI-SQUARE = 1739.231006 DOF = 403
disk+bulge:total_mass = 11.27124049
disk+bulge:r_eff_disk = 12.16817228
halo:fdm = 0.06921169832
dispprof:sigma0 = 29.36534085
geom:inc = 72
geom:pa = 145.5474635
geom:xshift = 0.0012542433
geom:yshift = -0.1281740091
geom:vel_shift = 25.16393383
INFO:DysmalPy:Iter 19 CHI-SQUARE = 1739.16602 DOF = 403
disk+bulge:total_mass = 11.27116728
disk+bulge:r_eff_disk = 12.16748897
halo:fdm = 0.06849834081
dispprof:sigma0 = 29.39382619
geom:inc = 72
geom:pa = 145.5310079
geom:xshift = 0.001254262134
geom:yshift = -0.1281740093
geom:vel_shift = 25.16345192
INFO:DysmalPy:Iter 20 CHI-SQUARE = 1739.088768 DOF = 403
disk+bulge:total_mass = 11.27103747
disk+bulge:r_eff_disk = 12.16351352
halo:fdm = 0.06834328557
dispprof:sigma0 = 29.48910938
geom:inc = 72
geom:pa = 145.5112834
geom:xshift = 0.001254247263
geom:yshift = -0.12817401
geom:vel_shift = 25.16981092
INFO:DysmalPy:Iter 21 CHI-SQUARE = 1739.068692 DOF = 403
disk+bulge:total_mass = 11.27103113
disk+bulge:r_eff_disk = 12.1637523
halo:fdm = 0.06841855337
dispprof:sigma0 = 29.56525146
geom:inc = 72
geom:pa = 145.5131879
geom:xshift = 0.001254248468
geom:yshift = -0.12817401
geom:vel_shift = 25.17058334
INFO:DysmalPy:Iter 22 CHI-SQUARE = 1739.068121 DOF = 403
disk+bulge:total_mass = 11.27101952
disk+bulge:r_eff_disk = 12.16317579
halo:fdm = 0.06840690692
dispprof:sigma0 = 29.52972761
geom:inc = 72
geom:pa = 145.5064353
geom:xshift = 0.001254248505
geom:yshift = -0.12817401
geom:vel_shift = 25.17074722
INFO:DysmalPy:
End: 2024-05-02 10:48:07.465394
******************
Time= 12.15 (sec), 0:12.15 (m:s)
MPFIT Status = 2
MPFIT Error/Warning Message = None
******************
Examine MPFIT results
Result plots
# Look at best-fit:
filepath = outdir_mpfit+"GS4_43501_mpfit_bestfit_halpha_2D.{}".format(plot_type)
Image(filename=filepath, width=600)

Directly generating result plots
Reload the galaxy, results files:
f_galmodel = outdir_mpfit + 'GS4_43501_model.pickle'
f_mpfit_results = outdir_mpfit + 'GS4_43501_mpfit_results.pickle'
gal, mpfit_results = fitting.reload_all_fitting(filename_galmodel=f_galmodel,
filename_results=f_mpfit_results, fit_method='mpfit')
Plot the best-fit results:
mpfit_results.plot_results(gal)

Result reports
We now look at the results reports, which include the best-fit values and uncertainties (as well as other fitting settings and output).
# Print report
print(mpfit_results.results_report(gal=gal))
###############################
Fitting for GS4_43501
Date: 2024-05-02 10:48:08.695418
obs: halpha_2D
Datafiles:
fit_velocity: True
fit_dispersion: True
fit_flux: False
moment: False
n_wholepix_z_min: 3
oversample: 1
oversize: 1
Fitting method: MPFIT
fit status: 2
pressure_support: True
pressure_support_type: 1
###############################
Fitting results
-----------
disk+bulge
total_mass 11.2710 +/- 0.0105
r_eff_disk 12.1632 +/- 0.3596
n_disk 1.0000 [FIXED]
r_eff_bulge 1.0000 [FIXED]
n_bulge 4.0000 [FIXED]
bt 0.3000 [FIXED]
mass_to_light 1.0000 [FIXED]
noord_flat True
-----------
halo
fdm 0.0684 +/- 0.0017
mvirial 10.3144 [TIED]
conc 5.0000 [FIXED]
-----------
dispprof
sigma0 29.5297 +/- 1.0188
-----------
zheightgaus
sigmaz 2.0661 [TIED]
-----------
geom
inc 72.0000 +/- 0.0000
pa 145.5064 +/- 0.3302
xshift 0.0013 +/- 0.0001
yshift -0.1282 +/- 0.0056
vel_shift 25.1707 +/- 0.4656
-----------
Adiabatic contraction: False
-----------
Red. chisq: 4.3153
-----------
obs halpha_2D: Rout,max,2D: 10.3258
To directly save the results report to a file, we can use the following:
(Note: by default, report files are saved as part of the fitting process)
# Save report to file:
f_mpfit_report = outdir_mpfit + 'mpfit_fit_report.txt'
mpfit_results.results_report(gal=gal, filename=f_mpfit_report)
4) MCMC Fitting
Get a clean copy of model, obs
gal = galaxy.Galaxy(z=1.613, name='GS4_43501')
obscopy = observation.Observation(name='halpha_2D', tracer='halpha')
obscopy.data = copy.deepcopy(data2d)
obscopy.instrument = copy.deepcopy(inst)
obscopy.mod_options = copy.deepcopy(obs.mod_options)
obscopy.fit_options = copy.deepcopy(obs.fit_options)
gal.add_observation(obscopy)
gal.model = copy.deepcopy(mod_set)
Set up MCMCFitter fitter with fitting parameters
# Options passed to emcee
## SHORT TEST:
nWalkers = 20
nCPUs = 4
scale_param_a = 5 #3
# The walkers were not exploring the parameter space well with scale_param_a = 3,
# but were getting 'stuck'. So to improve the walker movement with larger
# 'stretch' in the steps, we increased scale_param_a.
nBurn = 2
nSteps = 5
minAF = None
maxAF = None
nEff = 10
blob_name = 'mvirial' # Also save 'blob' values of Mvirial, calculated at every chain step
fitter = fitting.MCMCFitter(nWalkers=nWalkers, nCPUs=nCPUs,
scale_param_a=scale_param_a, nBurn=nBurn, nSteps=nSteps,
minAF=minAF, maxAF=maxAF, nEff=nEff, blob_name=blob_name)
Set up fit/plot output options
# Output directory
outdir_mcmc = outdir + 'MCMC/'
# Output options:
do_plotting = True
plot_type = 'png'
overwrite = True
output_options = config.OutputOptions(outdir=outdir_mcmc,
do_plotting=do_plotting,
plot_type=plot_type,
overwrite=overwrite)
Run Dysmalpy fitting: MCMC
mcmc_results = fitter.fit(gal, output_options)
INFO:DysmalPy:*************************************
INFO:DysmalPy: Fitting: GS4_43501 with MCMC
INFO:DysmalPy: obs: halpha_2D
INFO:DysmalPy: nSubpixels: 1
INFO:DysmalPy:
nCPUs: 4
INFO:DysmalPy:nWalkers: 20
INFO:DysmalPy:lnlike: oversampled_chisq=True
INFO:DysmalPy:
blobs: mvirial
INFO:DysmalPy:
mvirial_tied: <function tie_lmvirial_NFW at 0x176a55da0>
INFO:DysmalPy:
Burn-in:
Start: 2024-05-02 10:48:08.778789
INFO:DysmalPy: k=0, time.time=2024-05-02 10:48:08.779474, a_frac=nan
INFO:DysmalPy: k=1, time.time=2024-05-02 10:48:09.729780, a_frac=0.25
INFO:DysmalPy:
End: 2024-05-02 10:48:10.247523
******************
nCPU, nParam, nWalker, nBurn = 4, 9, 20, 2
Scale param a= 5
Time= 1.47 (sec), 0:1.47 (m:s)
Mean acceptance fraction: 0.225
Ideal acceptance frac: 0.2 - 0.5
Autocorr est: [nan nan nan nan nan nan nan nan nan]
******************
INFO:DysmalPy:
Ensemble sampling:
Start: 2024-05-02 10:48:10.973361
INFO:DysmalPy:ii=0, a_frac=0.2 time.time()=2024-05-02 10:48:11.308325
INFO:DysmalPy:0: acor_time =[nan nan nan nan nan nan nan nan nan]
INFO:DysmalPy:ii=1, a_frac=0.175 time.time()=2024-05-02 10:48:11.514939
INFO:DysmalPy:1: acor_time =[nan nan nan nan nan nan nan nan nan]
INFO:DysmalPy:ii=2, a_frac=0.18333333333333335 time.time()=2024-05-02 10:48:12.045867
INFO:DysmalPy:2: acor_time =[nan nan nan nan nan nan nan nan nan]
INFO:DysmalPy:ii=3, a_frac=0.2125 time.time()=2024-05-02 10:48:12.293471
INFO:DysmalPy:3: acor_time =[nan nan nan nan nan nan nan nan nan]
INFO:DysmalPy:ii=4, a_frac=0.22000000000000006 time.time()=2024-05-02 10:48:12.597950
INFO:DysmalPy:4: acor_time =[nan nan nan nan nan nan nan nan nan]
INFO:DysmalPy:Finished 5 steps
INFO:DysmalPy:
End: 2024-05-02 10:48:12.608378
******************
nCPU, nParam, nWalker, nSteps = 4, 9, 20, 5
Scale param a= 5
Time= 1.63 (sec), 0:1.63 (m:s)
Mean acceptance fraction: 0.220
Ideal acceptance frac: 0.2 - 0.5
Autocorr est: [nan nan nan nan nan nan nan nan nan]
******************
WARNING:root:Too few points to create valid contours
WARNING:root:Too few points to create valid contours
WARNING:root:Too few points to create valid contours
WARNING:root:Too few points to create valid contours
Examine MCMC results
Of course this (very short!) example looks terrible, but it’s instructive to see what’s happening even if you only did a very short / few walker MCMC test:
Trace
The individual walkers should move around in the parameter space over the chain iterations (not necessarily for every step; but there should be some exploration of the space)
# Look at trace:
filepath = outdir_mcmc+"GS4_43501_mcmc_trace.{}".format(plot_type)
Image(filepath, width=600)
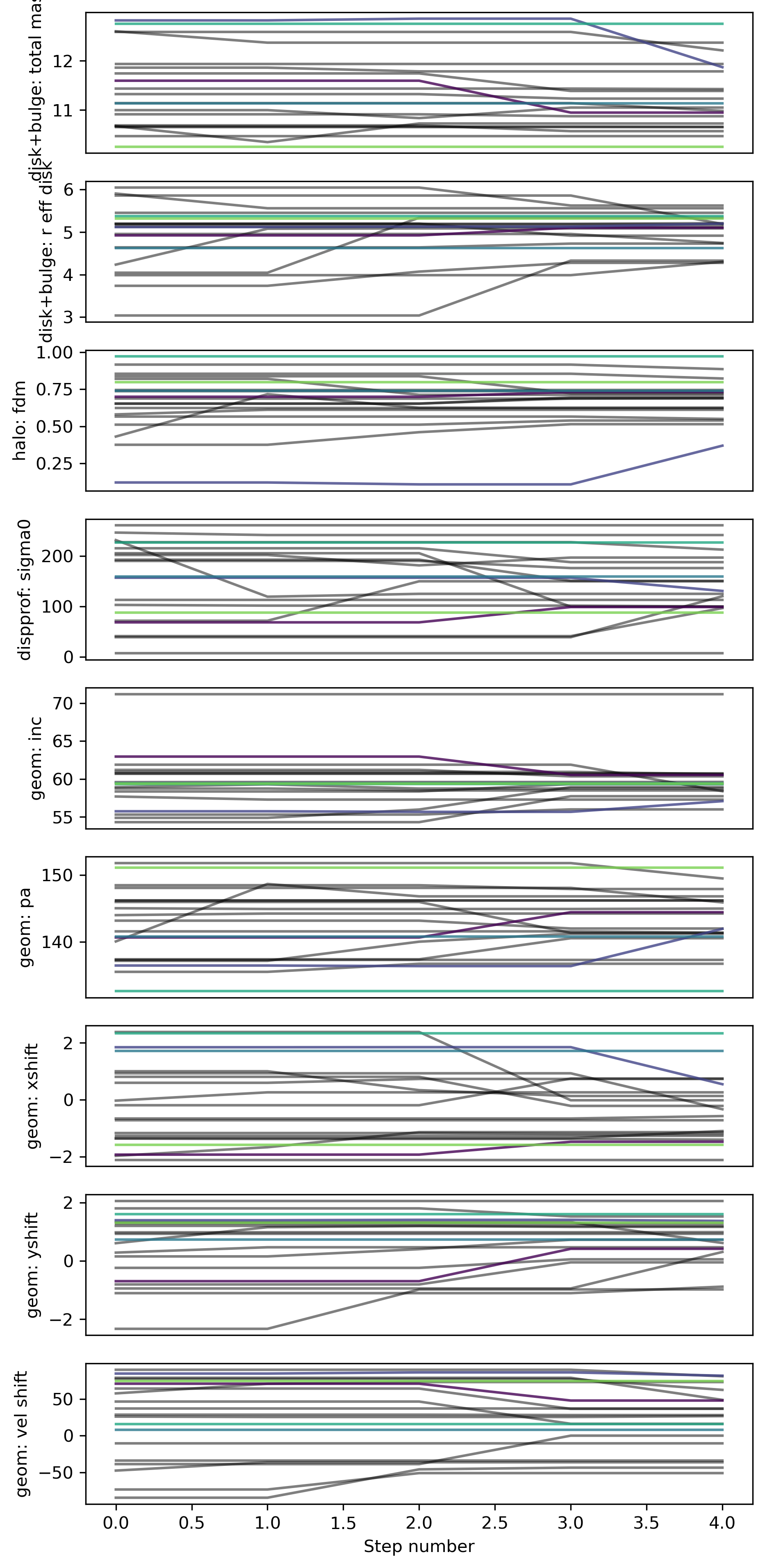
Best-fit
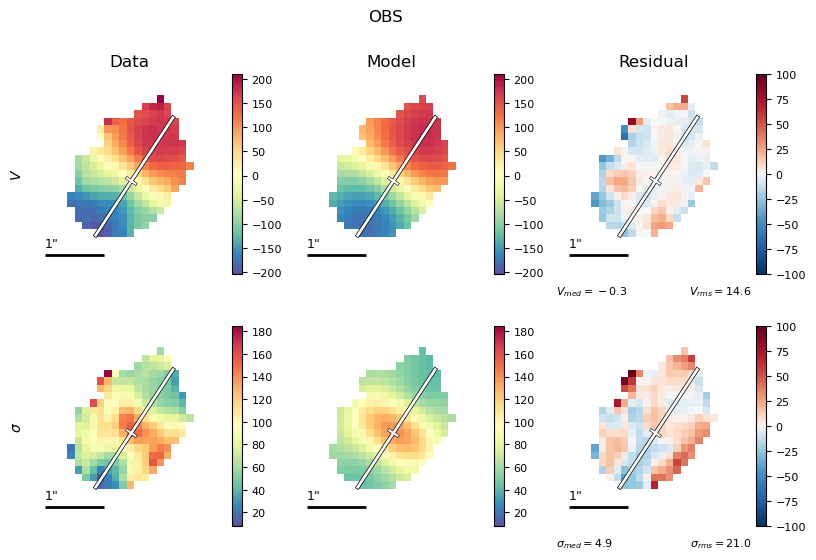
This is a good opportunity to check that the model PA and slit PA are correct, or else the data and model curves will have opposite shapes!
Also, it’s helpful to check that your data masking is reasonable.
Finally, this is a worthwhile chance to see if your “nuisance” geometry and spectral parameters (especially
xshift,yshift,vel_shifthave reasonable values, and if appropriate, reasonable bounds and priors.)
# Look at best-fit:
filepath = outdir_mcmc+"GS4_43501_mcmc_bestfit_halpha_2D.{}".format(plot_type)
Image(filepath, width=600)

Sampler “corner” plot
The “best-fit” MAP (by default taken to be the peak of each marginalized parameter posterior, independent of the other parameters) is marked with the solid blue line.
However, the MAP can also be found by jointly analyzing two or more parameters’ posterior space.
(→ see the example in the :ref:
1D example fit <dysmalpy_example_fit_1D.ipynb>tutorial)
Check to see that your Gaussian prior centers are marked in orange in the appropriate rows/columns (if any Gaussian priors are used).
The vertical dashed black lines show the 2.275%, 15.865%, 84.135%, 97.725% percentile intervals for the marginalized posterior for each parameter.
The vertical dashed purple lines show the shortest \(1\sigma\) interval, determined from the marginalized posterior for each parameter independently.
# Look at corner:
filepath = outdir_mcmc+"GS4_43501_mcmc_param_corner.{}".format(plot_type)
Image(filepath, height=620)
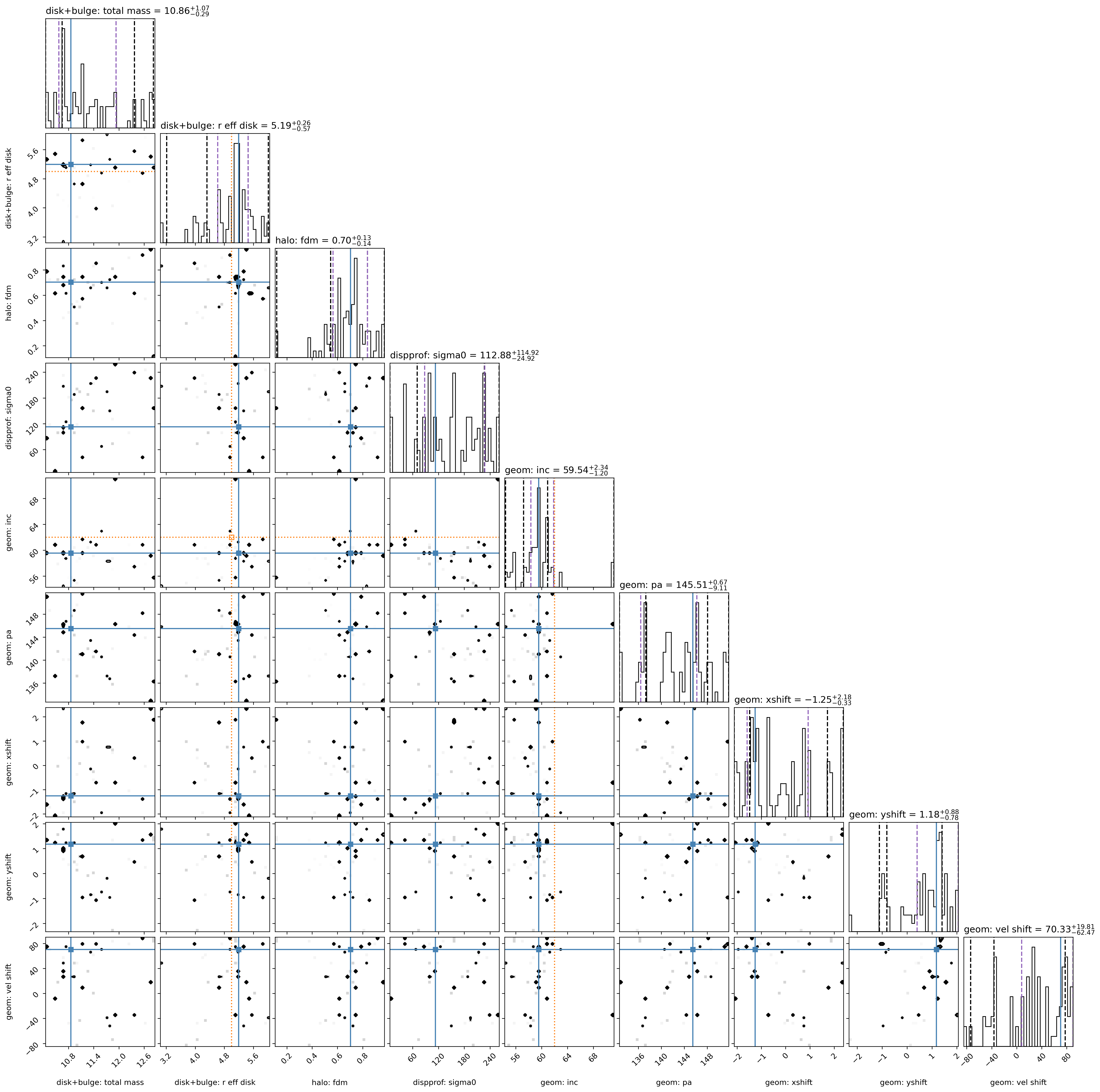
5) Visualize a real MCMC example
In the interest of time, let’s look at some results calculated previously, using 1000 walkers, 175 burn-in steps, and 200 steps.
Using 190 threads, it took about 21 minutes to run the MCMC fit.
First, you need to download the sampler from:
https://www.mpe.mpg.de/resources/IR/DYSMALPY/dysmalpy_optional_extra_files/JUPYTER_OUTPUT_2D.tar.gz
Decompress it in your preferred path (e.g.: /path/to/your/directory) and then set an environment variable to access that directory from this notebook. You can do that from the terminal running:
export DATADIR=/path/to/your/directory
Or, you can simply run the bash command from within the notebook as:
%env FULL_MCMC_RUNS_DATADIR=/path/to/your/directory
# A working example using the path of one of the maintainers:
%env FULL_MCMC_RUNS_DATADIR=/Users/jespejo/Dropbox/dysmalpy_example_files/JUPYTER_OUTPUT_2D/
env: FULL_MCMC_RUNS_DATADIR=/Users/jespejo/Dropbox/dysmalpy_example_files/JUPYTER_OUTPUT_2D/
# Now, add the environment variable to retrieve the data
_full_mcmc_runs = os.getenv('FULL_MCMC_RUNS_DATADIR')
Note
We used moment extraction for the model 2D profile extraction, as this does not seem to change the result much compared to gaussian extraction (as the observed maps were derived). This is likely because the extraction is over single pixels, so there is less complex line shape mixing as found in 1D extractions.
If this fit is done using a gaussian extraction, exactly following the data extraction method, it is generally slower.
When performing fits using moment extraction on data that was gaussian extracted, it is worth considering whether this is a reasonable thing to be doing. (Or at least check the final results with a gaussian extraction to see if there are systematic differences between the moment and gaussian maps.)
outdir_mcmc_full = _full_mcmc_runs + '/MCMC_full_run_nw1000_ns200_a5/'
Examine results:
Helpful for:
replotting
reanalyzing chain (eg, jointly constraining some posteriors)
Reload the galaxy, results files:
f_galmodel = outdir_mcmc_full + 'galaxy_model.pickle'
f_mcmc_results = outdir_mcmc_full + 'mcmc_results.pickle'
#----------------------------------------
## Fix module import
import sys
from dysmalpy import fitting_wrappers
sys.modules['fitting_wrappers'] = fitting_wrappers
#----------------------------------------
if os.path.isfile(f_galmodel) & os.path.isfile(f_mcmc_results):
gal_full, mcmc_results = fitting.reload_all_fitting(filename_galmodel=f_galmodel,
filename_results=f_mcmc_results, fit_method='mcmc')
else:
gal_full = mcmc_results = None
If necessary, also reload the sampler chain:
f_sampler = outdir_mcmc_full + 'mcmc_sampler.{}'.format(ftype_sampler)
if os.path.isfile(f_sampler) & (mcmc_results is not None):
mcmc_results.reload_sampler_results(filename=f_sampler)
WARNING:emcee.autocorr:The chain is shorter than 10 times the integrated autocorrelation time for 1 parameter(s). Use this estimate with caution and run a longer chain!
N/10 = 20;
tau: [19.6537326 19.9891195 19.55235249 19.98314067 19.7486466 19.41789169
19.19654616 20.31377879 19.68237055]
Plot the best-fit results:
if (gal_full is not None) & (mcmc_results is not None):
mcmc_results.plot_results(gal_full)
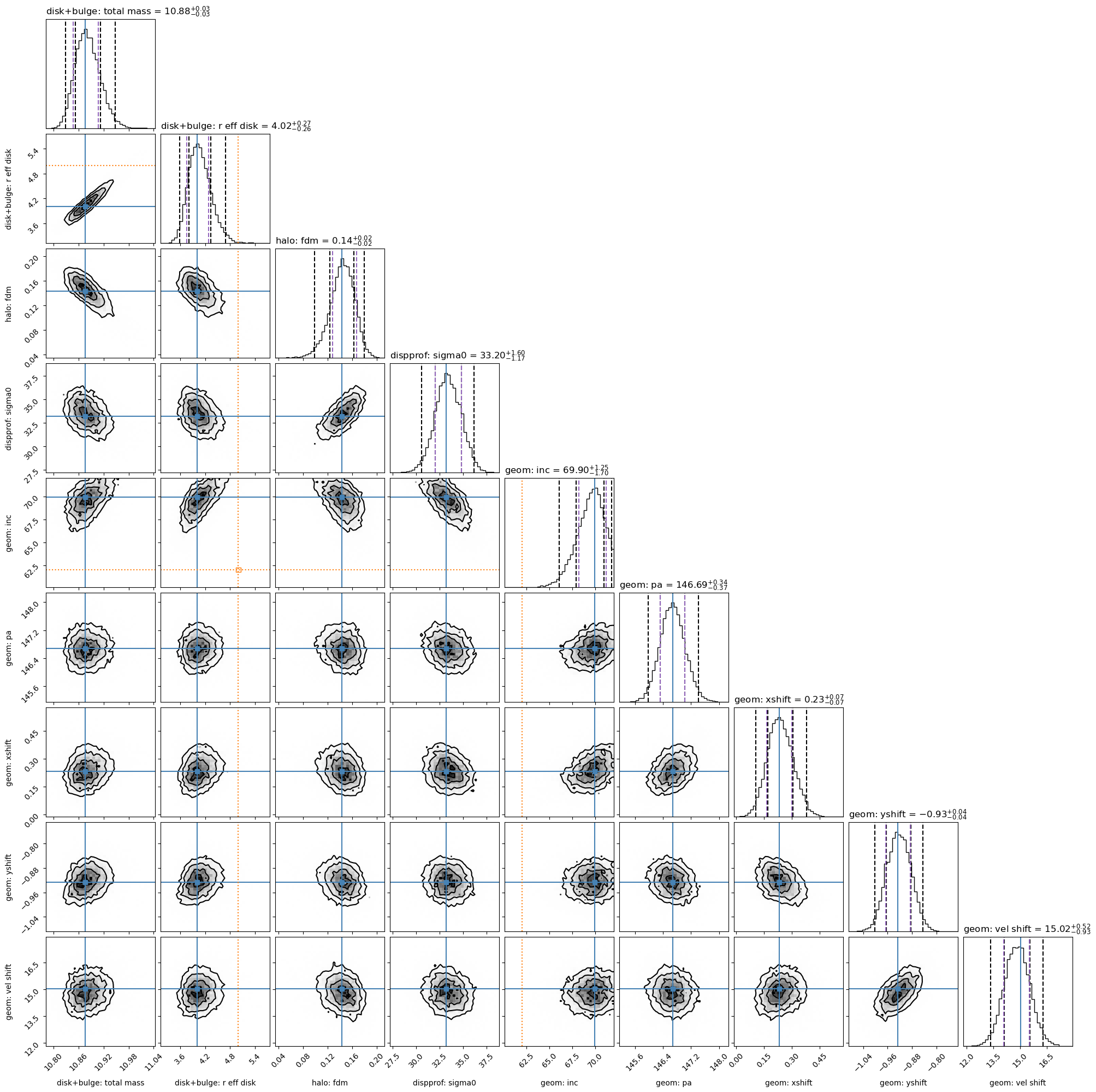
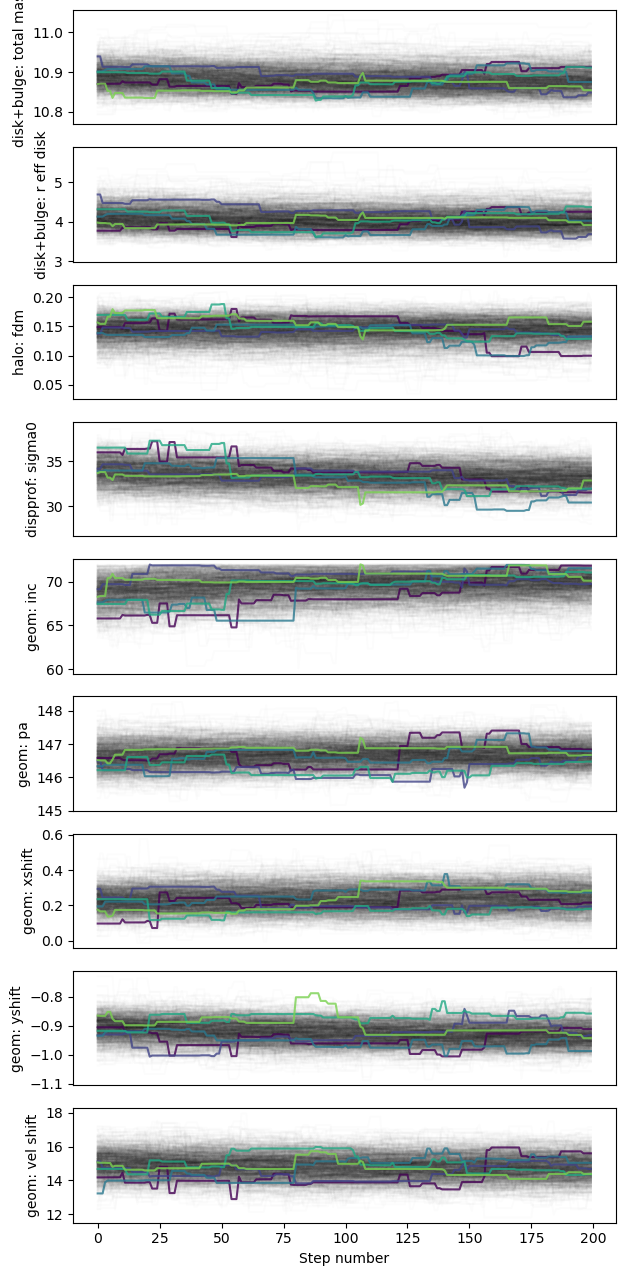
Results report:
# Print report
if (gal_full is not None) & (mcmc_results is not None):
print(mcmc_results.results_report(gal=gal_full))
###############################
Fitting for GS4_43501
Date: 2024-05-02 10:48:21.959682
obs: OBS
Datafiles:
fit_velocity: True
fit_dispersion: True
fit_flux: False
moment: True
n_wholepix_z_min: 3
oversample: 1
oversize: 1
Fitting method: MCMC
pressure_support: True
pressure_support_type: 1
###############################
Fitting results
-----------
disk+bulge
total_mass 10.8755 - 0.0278 + 0.0312
r_eff_disk 4.0151 - 0.2608 + 0.2707
n_disk 1.0000 [FIXED]
r_eff_bulge 1.0000 [FIXED]
n_bulge 4.0000 [FIXED]
bt 0.3000 [FIXED]
noord_flat True
-----------
halo
fdm 0.1433 - 0.0150 + 0.0242
mvirial 11.4323 [TIED]
conc 5.0000 [FIXED]
-----------
dispprof
sigma0 33.2030 - 1.1670 + 1.6011
-----------
zheightgaus
sigmaz 0.6820 [TIED]
-----------
geom
inc 69.8972 - 1.6953 + 1.2540
pa 146.6866 - 0.3726 + 0.3369
xshift 0.2318 - 0.0688 + 0.0688
yshift -0.9261 - 0.0410 + 0.0396
vel_shift 15.0223 - 0.9314 + 0.5174
-----------
mvirial 11.4323 - 0.1467 + 0.1957
-----------
Adiabatic contraction: False
-----------
Red. chisq: 2.3761
-----------
obs OBS: Rout,max,2D: 10.8553
Or save results report to file:
# Save report to file:
f_mcmc_report = outdir_mcmc + 'mcmc_fit_report.txt'
if (gal_full is not None) & (mcmc_results is not None):
mcmc_results.results_report(gal=gal_full, filename=f_mcmc_report)