Example Dysmalpy 3D fitting, using fitting wrapper
In this example, we use dysmalpy to measure the kinematics of galaxy GS4_43501 at \(z=1.613\) in 3D, using a fitting wrapper which simplifies the implementation of the fitting algorithm to the user. In this specific case, the fittign method is \(\texttt{MPFIT}\), as specified at the bottom of the fitting_3D_mpfit.params file. The notebook shows how to generate a mask on the cube and then find the best fit models for the velocity and velocity dispersion profiles. These fits allow us to measure quantities such as the total mass (disk+bulge), the effective radius \(r_\mathrm{eff}\), dark matter fraction \(f_\mathrm{DM}\) and velocity dispersion \(\sigma_0\).
The fitting includes the following components:
Disk + Bulge
NFW halo
Constant velocity dispersion
The structure of the notebook is the following:
Setup steps (and load the params file)
Generate mask for this IFU cube
Run
Dysmalpyfitting: 3D wrapper, with fit method= MPFITExamine results
Note
The results of 3D fitting can greatly depend on how the 3D cube is masked. An automated 3D mask generation script is included in DysmalPy (as shown in the example here), but care should be taken in choosing appropriate parameters. Furthermore, it may be benefitial to hand-edit the mask to include/exclude spaxels or spectral regions.
1) Setup steps
Import modules
from __future__ import (absolute_import, division, print_function,
unicode_literals)
from dysmalpy.fitting_wrappers import dysmalpy_fit_single
from dysmalpy.fitting_wrappers import utils_io
from dysmalpy import fitting, plotting
import os
import numpy as np
Setup notebook
# Setup plotting
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
mpl.rcParams['figure.dpi']= 100
mpl.rc("savefig", dpi=300)
from IPython.core.display import Image
Set data, output paths
Note this will override the datadir and outdir specified in the param file.
(This is useful for the example here. When running from command line, it’s recommended to properly set the directories in the param file.)
# Data directory (datadir = /YOUR/DATA/PATH/)
filepath = os.path.abspath(fitting.__file__)
datadir = os.sep.join(os.path.dirname(filepath).split(os.sep)[:-1]+["tests", "test_data", ""])
# Load the parameters file from the examples directory
param_path = os.sep.join(os.path.dirname(filepath).split(os.sep)[:-1]+["examples", "examples_param_files", ""])
param_filename = param_path+'fitting_3D_mpfit.params'
# Where to save output files (output = /YOUR/OUTPUTS/PATH)
outdir = '/Users/sedona/data/dysmalpy_test_examples/JUPYTER_OUTPUT_3D_FITTING_WRAPPER/'
outdir = '/Users/jespejo/Dropbox/Postdoc/Data/dysmalpy_test_examples/JUPYTER_OUTPUT_3D_FITTING_WRAPPER/'
outdir_mpfit = outdir + 'MPFIT/'
Settings in parameter file:
Note there are many commented out options / parameters. These given an more complete overview of the settings & parameters that can be specified with the fitting wrapper parameter files.
with open(param_filename, 'r') as f:
print(f.read())
# Example parameters file for fitting a single object with 1D data
# Note: DO NOT CHANGE THE NAMES IN THE 1ST COLUMN AND KEEP THE COMMAS!!
# See README for a description of each parameter and its available options.
# ******************************* OBJECT INFO **********************************
galID, GS4_43501 # Name of your object
z, 1.613 # Redshift
# ****************************** DATA INFO *************************************
datadir, None # Optional: Full path to data directory.
fdata_cube, gs4-43501_h250_21h30.fits.gz # Full path to vel map. Alternatively, just the filename if 'datadir' is set.
fdata_err, noise_gs4-43501_h250_21h30.fits.gz # Full path to vel. err map. Alternatively, just the filename if 'datadir' is set.
fdata_mask, GS4_43501_mask.fits # Full path to mask, if a separate mask is to be used.
# # -- strongly recommended to create a mask, or else use 'auto_gen_3D_mask, True' below
# Alternatively, just the filename if 'datadir' is set.
### FOR 3D, MUST SET CUBE, ETC by ** HAND **.
spec_orig_type, wave
spec_line_rest, 6564.
spec_line_rest_unit, angstrom
spec_vel_trim, -500. 500.
# l r b t
spatial_crop_trim, 37 68 34 65
xcenter, 51.75 #None # x/ycenter in UNCROPPED PIXELS. If None, assumed to be center of cube.
ycenter, 48.75 #None
data_inst_corr, False
auto_gen_3D_mask, True
auto_gen_mask_apply_skymask_first, True
auto_gen_mask_snr_thresh_pixel, None
auto_gen_mask_sig_segmap_thresh, 3.25
auto_gen_mask_npix_segmap_min, 5
auto_gen_mask_sky_var_thresh, 2. # H/J BAND # 3. # K/Y
auto_gen_mask_snr_int_flux_thresh, 3.
# ***************************** OUTPUT *****************************************
outdir, GS4_43501_3D_out/ # Full path for output directory
# ***************************** OBSERVATION SETUP ******************************
# Instrument Setup
# ------------------
pixscale, 0.125 # Pixel scale in arcsec/pixel
fov_npix, 37 # Number of pixels on a side of model cube
spec_type, velocity # DON'T CHANGE!
spec_start, -1000. # Starting value for spectral axis // generally don't change
spec_step, 10. # Step size for spectral axis in km/s // generally don't change
nspec, 201 # Number of spectral steps // generally don't change
# LSF Setup
# ---------
use_lsf, True # True/False if using an LSF
sig_inst_res, 51.0 # Instrumental dispersion in km/s
# PSF Setup
# ---------
psf_type, Gaussian # Gaussian, Moffat, or DoubleGaussian
psf_fwhm, 0.55 # PSF FWHM in arcsecs
psf_beta, -99. # Beta parameter for a Moffat PSF
# ## ELLIPTICAL PSF:
# psf_type, Gaussian # Gaussian, Moffat, or DoubleGaussian
# psf_fwhm_major, 0.55 # PSF major axis FWHM in arcsecs
# psf_fwhm_minor, 0.25 # PSF minor axis FWHM in arcsecs
# psf_PA, 0. # PA of PSF major axis, in deg E of N. (0=N, 90=E)
# psf_beta, -99. # Beta parameter for a Moffat PSF
# # DoubleGaussian: settings instead of psf_fwhm
# psf_type, DoubleGaussian
# psf_fwhm1, 0.16 # FWHM of PSF component 1, in arcsecs. SINFONI AO: 0.16
# psf_fwhm2, 0.48 # FWHM of PSF component 1, in arcsecs. SINFONI AO: 0.48
# psf_scale1, 0.368 # Flux scaling (*not* peak height) of component 1. SINFONI AO: 0.368
# psf_scale2, 0.632 # Flux scaling (*not* peak height) of component 2. SINFONI AO: 0.632
# **************************** SETUP MODEL *************************************
# Model Settings
# -------------
# List of components to use: SEPARATE WITH SPACES
## MUST always keep: geometry zheight_gaus
## RECOMMENDED: always keep: disk+bulge const_disp_prof
components_list, disk+bulge const_disp_prof geometry zheight_gaus halo
# possible options:
# disk+bulge, sersic, blackhole
# const_disp_prof, geometry, zheight_gaus, halo,
# radial_flow, uniform_planar_radial_flow, uniform_bar_flow, uniform_wedge_flow,
# unresolved_outflow, biconical_outflow,
# CAUTION: azimuthal_planar_radial_flow, variable_bar_flow, spiral_flow
# List of components that emit light. SEPARATE WITH SPACES
## Current options: disk+bulge / bulge / disk [corresponding to the mass disk+bulge component],
## also: light_sersic, light_gaussian_ring
light_components_list, disk
# NOTE: if a separate light profile (eg light_sersic) is used,
# this MUST be changed to e.g., 'light_components_list, light_sersic'
adiabatic_contract, False # Apply adiabatic contraction?
pressure_support, True # Apply assymmetric drift correction?
noord_flat, True # Apply Noordermeer flattenning?
oversample, 1 # Spatial oversample factor
oversize, 1 # Oversize factor
zcalc_truncate, True # Truncate in zgal direction when calculating or not
n_wholepix_z_min, 3 # Minimum number of whole pixels in zgal dir, if zcalc_truncate=True
# ********************************************************************************
# DISK + BULGE
# ------------
# Initial Values
total_mass, 11.0 # Total mass of disk and bulge log(Msun)
bt, 0.3 # Bulge-to-Total Ratio
r_eff_disk, 5.0 # Effective radius of disk in kpc
n_disk, 1.0 # Sersic index for disk
invq_disk, 5.0 # disk scale length to zheight ratio for disk
n_bulge, 4.0 # Sersic index for bulge
invq_bulge, 1.0 # disk scale length to zheight ratio for bulge
r_eff_bulge, 1.0 # Effective radius of bulge in kpc
# Fixed? True if its a fixed parameter, False otherwise
total_mass_fixed, False
r_eff_disk_fixed, False
bt_fixed, True
n_disk_fixed, True
r_eff_bulge_fixed, True
n_bulge_fixed, True
# Parameter bounds. Lower and upper bounds
total_mass_bounds, 10.0 13.0
bt_bounds, 0.0 1.0
r_eff_disk_bounds, 0.1 30.0
n_disk_bounds, 1.0 8.0
r_eff_bulge_bounds, 1.0 5.0
n_bulge_bounds, 1.0 8.0
# # ********************************************************************************
# # BLACK HOLE
# # ------------
#
# # Initial Values
# BH_mass, 11. # log(Msun)
#
# # Fixed? True if its a fixed parameter, False otherwise
# BH_mass_fixed, False
#
# # Parameter bounds. Lower and upper bounds
# BH_mass_bounds, 6. 18.
# # ********************************************************************************
# # Separate light profile: (Truncated) Sersic profile
# # ------------
# # Initial values
# L_tot_sersic, 1. # arbitrary units
# lr_eff, 4. # kpc
# lsersic_n, 1. # Sersic index of light profile
# lsersic_rinner, 0. # [kpc] Inner truncation radius of sersic profile. 0 = no truncation
# lsersic_router, inf # [kpc] Outer truncation radius of sersic profile. inf = no truncation
#
# # Fixed? True if its a fixed parameter, False otherwise
# L_tot_sersic_fixed, True
# lr_eff_fixed, False
# lsersic_n_fixed, True
# lsersic_rinner_fixed, True
# lsersic_router_fixed, True
#
# # Parameter bounds. Lower and upper bounds
# L_tot_sersic_bounds, 0. 2. # arbitrary units
# lr_eff_bounds, 0.5 15. # kpc
# lsersic_n_bounds, 0.5 8.
# lsersic_rinner_bounds, 0. 5. # kpc
# lsersic_router_bounds, 4. 20. # kpc
# # ********************************************************************************
# # Separate light profile: Gaussian ring
# # ------------
# # Initial values
# L_tot_gaus_ring, 1. # arbitrary units
# R_peak_gaus_ring, 6. # kpc
# FWHM_gaus_ring, 1. # kpc
#
# # Fixed? True if its a fixed parameter, False otherwise
# L_tot_gaus_ring_fixed, True
# R_peak_gaus_ring_fixed, True
# FWHM_gaus_ring_fixed, True
#
# # Parameter bounds. Lower and upper bounds
# L_tot_gaus_ring_bounds, 0. 2. # arbitrary units
# R_peak_gaus_ring_bounds, 0. 15. # kpc
# FWHM_gaus_ring_bounds, 0.1 10. # kpc
# ********************************************************************************
# ********************************************************************************
# ********************************************************************************
# DARK MATTER HALO
# ----------------
# Halo type: options: NFW / twopowerhalo / burkert / einasto / dekelzhao
halo_profile_type, NFW
# ** NOTE **: Uncomment the section below corresponding to the selected halo type.
# ********************************************************************************
# NFW halo
# Initial Values
mvirial, 11.5 # Halo virial mass in log(Msun)
halo_conc, 5.0 # Halo concentration parameter
fdm, 0.5 # Dark matter fraction at r_eff_disk
# Fixed? True if its a fixed parameter, False otherwise. Also set False if it will be tied (below)
mvirial_fixed, False
halo_conc_fixed, True
fdm_fixed, False
# Parameter bounds. Lower and upper bounds
mvirial_bounds, 10.0 13.0
halo_conc_bounds, 1.0 20.0
fdm_bounds, 0.0 1.0
# Tie the parameters?
fdm_tied, True # for NFW, fdm_tied=True determines fDM from Mvirial (+baryons)
mvirial_tied, False # for NFW, mvirial_tied=True determines Mvirial from fDM (+baryons)
# ********************************************************************************
# # ********************************************************************************
# # Two-power halo
#
# # Initial Values
# mvirial, 11.5 # Halo virial mass in log(Msun)
# halo_conc, 5.0 # Halo concentration parameter
# fdm, 0.5 # Dark matter fraction at r_eff_disk
# alpha, 1. # TPH: inner slope. NFW has alpha=1
# beta, 3. # TPH: outer slope. NFW has beta=3
#
# # Fixed? True if its a fixed parameter, False otherwise. Also set False if it will be tied (below)
# mvirial_fixed, False
# halo_conc_fixed, True
# fdm_fixed, False
# alpha_fixed, False
# beta_fixed, True
#
# # Parameter bounds. Lower and upper bounds
# mvirial_bounds, 10.0 13.0
# halo_conc_bounds, 1.0 20.0
# fdm_bounds, 0.0 1.0
# alpha_bounds, 0.0 3.0
# beta_bounds, 1.0 4.0
#
# # Tie the parameters?
# fdm_tied, True # for non-NFW, fdm_tied=True determines fDM from other halo params (+baryons)
# mvirial_tied, True # for non-NFW, mvirial_tied=True determines Mvirial from SMHM+fgas + baryon total_mass
# alpha_tied, False # for TPH, alpha_tied=True determines alpha from free fDM + other parameters.
#
# ### OTHER SETTINGS:
# mhalo_relation, Moster18 ## SMHM relation to use for tying Mvir to Mbar. options: Moster18 / Behroozi13
#
# fgas, 0.5 # Gas fraction for SMHM inference of Mvir if 'mvirial_tied=True'
# lmstar, -99. # Currently code uses fgas to infer lmstar
# # from fitting baryon total_mass for SMHM relation
# # ********************************************************************************
# # ********************************************************************************
# # Burkert halo
#
# # Initial Values
# mvirial, 11.5 # Halo virial mass in log(Msun)
# halo_conc, 5.0 # Halo concentration parameter
# fdm, 0.5 # Dark matter fraction at r_eff_disk
# rB, 10. # Burkert: Halo core radius, in kpc
#
# # Fixed? True if its a fixed parameter, False otherwise. Also set False if it will be tied (below)
# mvirial_fixed, False
# halo_conc_fixed, True
# fdm_fixed, False
# rB_fixed, False
#
# # Parameter bounds. Lower and upper bounds
# mvirial_bounds, 10.0 13.0
# halo_conc_bounds, 1.0 20.0
# fdm_bounds, 0.0 1.0
# rB_bounds, 1.0 20.0
#
# # Tie the parameters?
# fdm_tied, True # for non-NFW, fdm_tied=True determines fDM from other halo params (+baryons)
# mvirial_tied, True # for non-NFW, mvirial_tied=True determines Mvirial from SMHM+fgas + baryon total_mass
# rB_tied, False # for Burkert, rB_tied=True determines rB from free fDM + other parameters.
#
# ### OTHER SETTINGS:
# mhalo_relation, Moster18 ## SMHM relation to use for tying Mvir to Mbar. options: Moster18 / Behroozi13
#
# fgas, 0.5 # Gas fraction for SMHM inference of Mvir if 'mvirial_tied=True'
# lmstar, -99. # Currently code uses fgas to infer lmstar
# # from fitting baryon total_mass for SMHM relation
# # ********************************************************************************
# # ********************************************************************************
# # Einasto halo
# # Initial Values
# mvirial, 11.5 # Halo virial mass in log(Msun)
# halo_conc, 5.0 # Halo concentration parameter
# fdm, 0.5 # Dark matter fraction at r_eff_disk
# alphaEinasto, 1. # Einasto: Halo profile index
#
# # Fixed? True if its a fixed parameter, False otherwise. Also set False if it will be tied (below)
# mvirial_fixed, False
# halo_conc_fixed, True
# fdm_fixed, False
# alphaEinasto_fixed, False
#
# # Parameter bounds. Lower and upper bounds
# mvirial_bounds, 10.0 13.0
# halo_conc_bounds, 1.0 20.0
# fdm_bounds, 0.0 1.0
# alphaEinasto_bounds, 0.0 2.0
#
# # Tie the parameters?
# fdm_tied, True # for non-NFW, fdm_tied=True determines fDM from other halo params (+baryons)
# mvirial_tied, True # for non-NFW, mvirial_tied=True determines Mvirial from SMHM+fgas + baryon total_mass
# alphaEinasto_tied, False # for Einasto, alphaEinasto_tied=True determines alphaEinasto from free fDM + other params.
#
# ### OTHER SETTINGS:
# mhalo_relation, Moster18 ## SMHM relation to use for tying Mvir to Mbar. options: Moster18 / Behroozi13
#
# fgas, 0.5 # Gas fraction for SMHM inference of Mvir if 'mvirial_tied=True'
# lmstar, -99. # Currently code uses fgas to infer lmstar
# # from fitting baryon total_mass for SMHM relation
# # ********************************************************************************
# # ********************************************************************************
# # Dekel-Zhao halo
# # Initial Values
# mvirial, 12.0 # Halo virial mass in log(Msun)
# s1, 1.5 # Inner logarithmic slope (at resolution r1=0.01*Rvir)
# c2, 25.0 # Concentration parameter (defined relative to c, a)
# fdm, 0.5 # Dark matter fraction at r_eff_disk
#
# # Fixed? True if its a fixed parameter, False otherwise. Also set False if it will be tied (below)
# mvirial_fixed, False
# s1_fixed, False
# c2_fixed, False
# fdm_fixed, False
#
# # Parameter bounds. Lower and upper bounds
# mvirial_bounds, 10.0 13.0 # log(Msun)
# s1_bounds, 0.0 2.0
# c2_bounds, 0.0 40.0
# fdm_bounds, 0.0 1.0
#
# # Tie the parameters?
# mvirial_tied, True # mvirial_tied=True determines Mvirial from fDM, s1, c2.
# s1_tied, True # Tie the s1 to M*/Mvir using best-fit Freundlich+20 (Eqs 45, 47, 48, Table 1)
# c2_tied, True # Tie the c2 to M*/Mvir using best-fit Freundlich+20 (Eqs 47, 49, Table 1)
# fdm_tied, False # for non-NFW, fdm_tied=True determines fDM from other halo params (+baryons)
#
# ### OTHER SETTINGS:
# lmstar, 10.5 # Used to infer s1, c2 if s1_tied or c2_tied = True
#
# # ********************************************************************************
# ********************************************************************************
# INTRINSIC DISPERSION PROFILE
# ------------------
# Initial Values
sigma0, 39.0 # Constant intrinsic dispersion value
# Fixed? True if its a fixed parameter, False otherwise
sigma0_fixed, False
# Parameter bounds. Lower and upper bounds
sigma0_bounds, 5.0 300.0
# ********************************************************************************
# ********************************************************************************
# ********************************************************************************
# HIGHER ORDER COMPONENTS: INFLOW, OUTFLOW
# ----------------------------------------
# # ********************************************************************************
# # UNIFORM SPHERICAL RADIAL FLOW -- in rhat direction in spherical coordinates
# # radial_flow
# # -------------------
#
# vr, -90. # Radial flow [km/s]. Positive: Outflow. Negative: Inflow.
# # ********************************************************************************
# # UNIFORM PLANAR RADIAL FLOW -- in Rhat direction in cylindrical coordinates
# # (eg, radial in galaxy midplane)
# # uniform_planar_radial_flow
# # -------------------
#
# vr, -90. # Radial flow [km/s]. Positive: Outflow. Negative: Inflow.
# # ********************************************************************************
# # UNIFORM BAR FLOW -- in xhat direction along bar in cartesian coordinates,
# # with bar at an angle relative to galaxy major axis (blue)
# # uniform_bar_flow
# # -------------------
#
# vbar, -90. # Bar flow [km/s]. Positive: Outflow. Negative: Inflow.
# phi, 90. # Azimuthal angle of bar [degrees], counter-clockwise from blue major axis.
# # Default is 90 (eg, along galaxy minor axis)
# bar_width, 2 # Width of the bar perpendicular to bar direction.
# # Bar velocity only is nonzero between -bar_width/2 < ygal < bar_width/2.
# # ********************************************************************************
# # UNIFORM WEDGE FLOW -- in planar radial flow in cylindrical coordinates, restricted to pos, neg wedges
# # uniform_wedge_flow
# # -------------------
#
# vr, -90. # Radial flow [km/s]. Positive: Outflow. Negative: Inflow.
# theta, 60. # Opening angle of wedge [deg]. (the full angular span)
# phi, 90. # Angle offset relative to the galaxy angle, so the wedge center is at phi.
# # Default: 90 deg, so centered along minor axis
# # ********************************************************************************
# # UNRESOLVED OUTFLOW -- at galaxy center (ie, AGN unresolved outflow)
# # unresolved_outflow
# # -------------------
#
# vcenter, 0. # Central velocity of the Gaussian in km/s
# fwhm, 1000. # FWHM of the Gaussian in km/s
# amplitude, 1.e12 # Amplitude of the Gaussian, for flux in ~M/L=1 luminosity units
# # with the dimming applied ... roughly ....
# # ********************************************************************************
# # BICONICAL OUTFLOW
# # biconical_outflow
# # -------------------
#
# n, 0.5 # Power law index
# vmax, 500. # Maximum velocity of the outflow in km/s
# rturn, 5. # Turn-over radius in kpc of the velocty profile
# thetain, 30. # Half inner opening angle in degrees. Measured from the bicone axis
# dtheta, 20. # Difference between inner and outer opening angle in degrees
# rend, 10. # Maximum radius of the outflow in kpc
# norm_flux, 8. # Log flux amplitude of the outflow at r = 0.
# # Need to check dimming/flux conventions
# tau_flux, 1. # Exponential decay rate of the flux
# biconical_profile_type, both # Type of velocity profile:
# # 'both', 'increase', 'decrease', 'constant'
# biconical_outflow_dispersion, 80. # Dispersion (stddev of gaussian) of biconical outflow, km/s
# # ********************************************************************************
# # VARIABLE BAR FLOW -- in xhat direction along bar in cartesian coordinates,
# # with bar at an angle relative to galaxy major axis (blue)
# # CAUTION!!!
# # variable_bar_flow
# # -------------------
#
# vbar_func_bar_flow, -90.*np.exp(-R/5.) # Bar flow FUNCTION [km/s]. Positive: Outflow. Negative: Inflow.
# phi, 90. # Azimuthal angle of bar [degrees], counter-clockwise from blue major axis.
# # Default is 90 (eg, along galaxy minor axis)
# bar_width, 2 # Width of the bar perpendicular to bar direction.
# # Bar velocity only is nonzero between -bar_width/2 < ygal < bar_width/2.
# # ********************************************************************************
# # AZIMUTHAL PLANAR RADIAL FLOW -- in Rhat direction in cylindrical coordinates
# # (eg, radial in galaxy midplane), with an added azimuthal term
# # CAUTION!!!
# # azimuthal_planar_radial_flow
# # -------------------
#
# vr_func_azimuthal_planar_flow, -90.*np.exp(-R/5.) # Radial flow [km/s].
# # Positive: Outflow. Negative: Inflow.
# m, 2 # Number of modes in the azimuthal pattern. m=0 gives a purely radial profile.
# phi0, 0. # Angle offset relative to the galaxy angle [deg],
# # so the azimuthal variation goes as cos(m(phi_gal - phi0))
# # ********************************************************************************
# # SPIRAL DENSIY WAVE FLOW -- as in Davies et al. 2009, ApJ, 702, 114
# # Here assuming CONSTANT velocity -- try to match real Vrot...
# # CAUTION!!! NO SPACES IN FUNCTION DEFINITONS!
# # spiral_flow
# # -------------------
#
# Vrot_func_spiral_flow, 150.+0.*R # Unperturbed rotation velocity of the galaxy
# dVrot_dR_func_spiral_flow, 0.*R # Derivative of Vrot(R) -- ideally evaluated analytically, otherwise very slow.
# rho0_func_spiral_flow, 1.e11*np.exp(-R/5.) # Unperturbed midplane density profile of the galaxy
# f_func_spiral_flow, (np.sqrt(m**2-2.)*Vrot(R)/cs)*np.log(R) # Function describing the spiral shape, m*phi = f(R)
# # with k = df/dR
# k_func_spiral_flow, (np.sqrt(m**2-2.)*Vrot(R)/cs)/R # Function for radial wavenumber
#
# m, 2 # Number of photometric/density spiral arms.
# cs, 10. # Sound speed of medium, in km/s.
# epsilon, 1. # Density contrast of perturbation (unitless).
# Om_p, 0. # Angular speed of the driving force, Omega_p
# phi0, 0. # Angle offset of the arm winding, in degrees. Default: 0.
# ********************************************************************************
# ********************************************************************************
# ********************************************************************************
# ********************************************************************************
# ZHEIGHT PROFILE
# ---------------
# Initial Values
sigmaz, 0.9 # Gaussian width of the galaxy in z, in kpc
# Fixed? True if its a fixed parameter, False otherwise
sigmaz_fixed, False
# Parameter bounds. Lower and upper bounds
sigmaz_bounds, 0.1 1.0
# Tie the zheight to the effective radius of the disk?
# If set to True, make sure sigmaz_fixed is False
zheight_tied, True
# GEOMETRY
# --------
# Initial Values
inc, 62. # Inclination of galaxy, 0=face-on, 90=edge-on
pa, 142.
xshift, 0. # pixels
yshift, 0. # pixels
vel_shift, 0. # km/s
# Fixed? True if its a fixed parameter, False otherwise
inc_fixed, False
pa_fixed, False
xshift_fixed, False
yshift_fixed, False
vel_shift_fixed, False
# Parameter bounds. Lower and upper bounds
inc_bounds, 42. 82.
pa_bounds, 132. 152.
xshift_bounds, -1.5 1.5 # pixels
yshift_bounds, -1.5 1.5 # pixels
vel_shift_bounds, -100. 100. # km/s
# **************************** Fitting Settings ********************************
fit_method, mpfit # mcmc, nested, or mpfit
do_plotting, True # Produce all output plots?
# *** Note ***: fitflux and fitdispersion are not supported for 3D,
# because these are implicitly included in the cube!
# MPFIT Settings
#----------------
maxiter, 200 # Maximum number of iterations before mpfit quits
Add some settings for this notebook example:
plot_type = 'png'
2) Generate mask for this IFU cube
As we don’t have a premade mask for the data cube, we will use an automated script to generate a mask.
This script can mask skylines (based on the spectral variance spectrum), as well as applying a segmentation map to limit the fitting to a contiguous region based on the integrated line flux.
Load parameters and galaxy
First, you need to download the test fits files from:
https://www.mpe.mpg.de/resources/IR/DYSMALPY/dysmalpy_optional_extra_files/gs4-43501_h250_21h30.fits.gz
and
https://www.mpe.mpg.de/resources/IR/DYSMALPY/dysmalpy_optional_extra_files/noise_gs4-43501_h250_21h30.fits.gz
You can either copy (or move) these files into your datadir directory, or you can create the envirnoment variables in the terminal as:
export FDATA_CUBE=/YOUR/DATA/PATH/gs4-43501_h250_21h30.fits.gz
export FDATA_ERR=/YOUR/DATA/PATH/noise_gs4-43501_h250_21h30.fits.gz
# You can also run the bash commands from within the notebook as (e.g. a directoy of one of the maintainers):
%env FDATA_CUBE=/Users/jespejo/Dropbox/Postdoc/dysmalpy/dysmalpy/dysmalpy/tests/test_data/gs4-43501_h250_21h30.fits.gz
%env FDATA_ERR=/Users/jespejo/Dropbox/Postdoc/dysmalpy/dysmalpy/dysmalpy/tests/test_data/noise_gs4-43501_h250_21h30.fits.gz
env: FDATA_CUBE=/Users/jespejo/Dropbox/Postdoc/dysmalpy/dysmalpy/dysmalpy/tests/test_data/gs4-43501_h250_21h30.fits.gz
env: FDATA_ERR=/Users/jespejo/Dropbox/Postdoc/dysmalpy/dysmalpy/dysmalpy/tests/test_data/noise_gs4-43501_h250_21h30.fits.gz
params = utils_io.read_fitting_params(fname=param_filename)
# check if the files are inside the datadir, if so load them from there:
if os.path.exists(datadir+"gs4-43501_h250_21h30.fits.gz") and os.path.exists(datadir+"noise_gs4-43501_h250_21h30.fits.gz"):
gal = utils_io.load_galaxy(params=params, datadir=datadir, skip_mask=True, skip_automask=True)
# if not, load them from the environment variables
else:
params["fdata_cube"] = os.getenv('FDATA_CUBE')
params["fdata_err"] = os.getenv('FDATA_ERR')
gal = utils_io.load_galaxy(params=params, datadir=None, skip_mask=True, skip_automask=True)
INFO:DysmalPy:dysmalpy.Galaxy:
********************************************************************
*** INFO ***
instrument.fov[0,1]=(37,37) is being reset to match 3D cube (31, 31)
********************************************************************
INFO:DysmalPy:dysmalpy.Galaxy:
********************************************************************
*** INFO ***
instrument spectral settings are being reset
(spec_type=velocity, spec_start=-1000.00 km / s, spec_step=10.00 km / s, nspec=201)
to match 3D cube
(spec_type=velocity, spec_start=-484.72 km / s, spec_step=34.08 km / s, nspec=29)
********************************************************************
Note that any mismatches in the instrument FOV or spectral settings have been automatically set, and reset to match the input data cube.
Generate mask
mask, mask_dict = utils_io.generate_3D_mask(obs=gal.observations['OBS'], params=params)
++++++++++++++++++++++++++++++++++++++++++++++++++++++
Creating 3D auto mask with the following settings:
sig_segmap_thresh = 3.25
npix_segmap_min = 5
snr_int_flux_thresh = 3
snr_thresh_pixel = None
sky_var_thresh = 2
apply_skymask_first = True
++++++++++++++++++++++++++++++++++++++++++++++++++++++
Masking segmap: used exclude_percentile=15.0
Note that we set the relevant parameters (sig_segmap_thresk, npix_segmap_min, sky_var_thresh, etc.) in the parameters file, by prepending auto_gen_mask_ to the keyword names.
These parameters (sig_segmap_thresk, npix_segmap_min, sky_var_thresh, etc.) can also be passed directly to utils_io.generate_3D_mask(), which can be helpful when exploring different parameter settings.
Examine mask info
First look at the integrated spatial flux maps and segmaps
plotting.plot_3D_data_automask_info(gal.observations['OBS'], mask_dict)
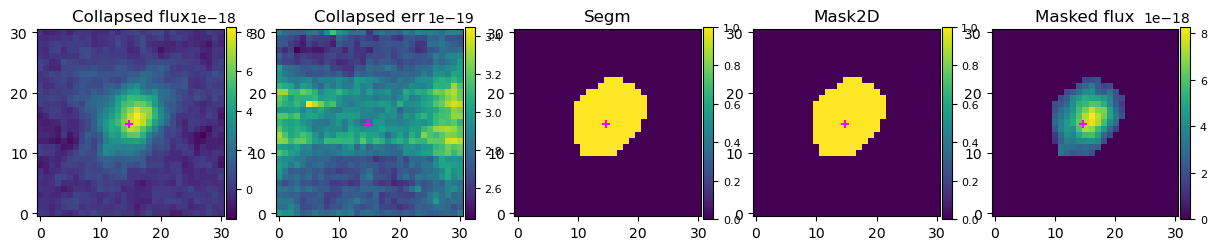
Then examine the per-spaxel spectra, to see if the skyline masking/velocity cropping and the segmentation map masking is sufficient.
gal.observations['OBS'].data.mask = np.array(mask, dtype=bool)
plotting.plot_spaxel_compare_3D_cubes(gal.observations['OBS'], skip_fits=True)
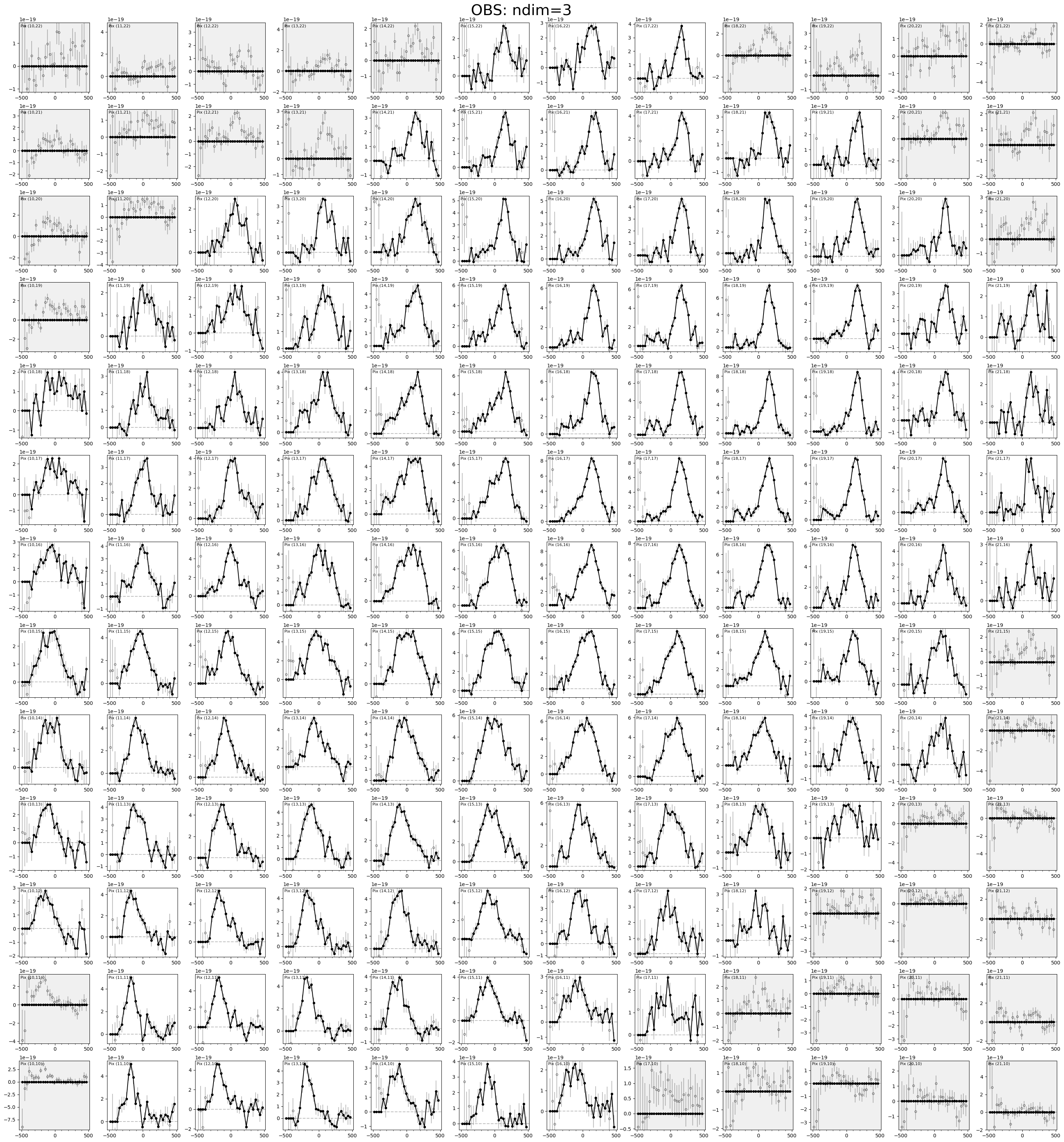
Save mask
After finalizing the parameters, save a copy of the mask to a FITS file (to the filename specified with fdata_mask in the params file).
Note we will backmap the mask to the uncropped cube size, so the raw data and error cubes can be directly compared to raw mask file. (The same cropping will be applied to the data, error, and mask cubes when loading the data for fitting.)
utils_io.save_3D_mask(obs=gal.observations['OBS'], mask=mask,
filename=datadir+params['fdata_mask'],
save_uncropped_size=True, overwrite=True)
Note that we set the relevant parameters (sig_segmap_thresk, npix_segmap_min, sky_var_thresh, etc.) in the parameters file.
If fdata_mask is not set in the param file, or if the file is not found, then when running the fitting wrapper the behavior will fall back to creating a mask on the fly (as by default skip_automask=False).
In this case, the automatic mask generation uses the specified automask parameters in the param file (or else falls back to the automask defaults, if not specified).
However, if the mask is generated on the fly, it will not be saved to a separate FITS file (though it will be accessible in the galaxy object stored in the “galaxy model” pickle).
For this example, we choose to directly save the mask to a FITS file, for clarity.
3) Run Dysmalpy fitting: 3D wrapper, with fit method= MPFIT
dysmalpy_fit_single.dysmalpy_fit_single(param_filename=param_filename,
datadir=datadir, outdir=outdir_mpfit,
plot_type=plot_type, overwrite=True)
INFO:DysmalPy:dysmalpy.Galaxy:
********************************************************************
*** INFO ***
instrument.fov[0,1]=(37,37) is being reset to match 3D cube (31, 31)
********************************************************************
INFO:DysmalPy:dysmalpy.Galaxy:
********************************************************************
*** INFO ***
instrument spectral settings are being reset
(spec_type=velocity, spec_start=-1000.00 km / s, spec_step=10.00 km / s, nspec=201)
to match 3D cube
(spec_type=velocity, spec_start=-484.72 km / s, spec_step=34.08 km / s, nspec=29)
********************************************************************
INFO:DysmalPy:*************************************
INFO:DysmalPy: Fitting: GS4_43501 using MPFIT
INFO:DysmalPy: obs: OBS
INFO:DysmalPy: nSubpixels: 1
INFO:DysmalPy: mvirial_tied: False
INFO:DysmalPy:
MPFIT Fitting:
Start: 2024-05-02 10:46:14.208246
INFO:DysmalPy:Iter 1 CHI-SQUARE = 4218.164951 DOF = 27860
disk+bulge:total_mass = 11
disk+bulge:r_eff_disk = 5
halo:mvirial = 11.5
dispprof_LINE:sigma0 = 39
geom_1:inc = 62
geom_1:pa = 142
geom_1:xshift = 0
geom_1:yshift = 0
geom_1:vel_shift = 0
INFO:DysmalPy:Iter 2 CHI-SQUARE = 3675.59407 DOF = 27860
disk+bulge:total_mass = 10.80433652
disk+bulge:r_eff_disk = 3.837666768
halo:mvirial = 11.30866673
dispprof_LINE:sigma0 = 59.37682188
geom_1:inc = 80.00155846
geom_1:pa = 146.705436
geom_1:xshift = -0.2735720121
geom_1:yshift = -0.2298953424
geom_1:vel_shift = -2.908511392
INFO:DysmalPy:Iter 3 CHI-SQUARE = 3222.803726 DOF = 27860
disk+bulge:total_mass = 10.8350294
disk+bulge:r_eff_disk = 3.920545563
halo:mvirial = 11.81621325
dispprof_LINE:sigma0 = 60.42119537
geom_1:inc = 74.47722512
geom_1:pa = 145.6248469
geom_1:xshift = -0.2200430516
geom_1:yshift = -0.2537370447
geom_1:vel_shift = -0.9221022035
INFO:DysmalPy:Iter 4 CHI-SQUARE = 3174.107713 DOF = 27860
disk+bulge:total_mass = 10.80162586
disk+bulge:r_eff_disk = 3.786452426
halo:mvirial = 12.66231385
dispprof_LINE:sigma0 = 69.54514214
geom_1:inc = 63.46311126
geom_1:pa = 145.7917685
geom_1:xshift = -0.03709494476
geom_1:yshift = -0.4743505199
geom_1:vel_shift = -2.439100695
INFO:DysmalPy:Iter 5 CHI-SQUARE = 3170.376884 DOF = 27860
disk+bulge:total_mass = 10.76828722
disk+bulge:r_eff_disk = 2.576580453
halo:mvirial = 12.86092446
dispprof_LINE:sigma0 = 70.03421873
geom_1:inc = 51.44811069
geom_1:pa = 147.4904322
geom_1:xshift = 0.03841825081
geom_1:yshift = -0.6161729455
geom_1:vel_shift = -5.049030366
INFO:DysmalPy:Iter 6 CHI-SQUARE = 3064.89553 DOF = 27860
disk+bulge:total_mass = 10.78224819
disk+bulge:r_eff_disk = 2.155514718
halo:mvirial = 12.91137608
dispprof_LINE:sigma0 = 63.78749875
geom_1:inc = 49.33641089
geom_1:pa = 147.4236358
geom_1:xshift = 0.1292068766
geom_1:yshift = -0.6318799829
geom_1:vel_shift = -4.314706328
INFO:DysmalPy:Iter 7 CHI-SQUARE = 3009.128776 DOF = 27860
disk+bulge:total_mass = 10.82206632
disk+bulge:r_eff_disk = 2.071378948
halo:mvirial = 12.83244043
dispprof_LINE:sigma0 = 60.05971527
geom_1:inc = 46.16818109
geom_1:pa = 147.4399653
geom_1:xshift = 0.08477367771
geom_1:yshift = -0.6291123471
geom_1:vel_shift = -5.710037106
INFO:DysmalPy:Iter 8 CHI-SQUARE = 3008.950957 DOF = 27860
disk+bulge:total_mass = 10.8781941
disk+bulge:r_eff_disk = 2.029116462
halo:mvirial = 12.8297734
dispprof_LINE:sigma0 = 59.40780632
geom_1:inc = 42
geom_1:pa = 147.5578397
geom_1:xshift = 0.1045898794
geom_1:yshift = -0.6423934605
geom_1:vel_shift = -5.232484049
INFO:DysmalPy:Iter 9 CHI-SQUARE = 3008.019322 DOF = 27860
disk+bulge:total_mass = 10.83070952
disk+bulge:r_eff_disk = 2.05294434
halo:mvirial = 12.79751151
dispprof_LINE:sigma0 = 59.22987554
geom_1:inc = 45.37229525
geom_1:pa = 147.6098562
geom_1:xshift = 0.1078168537
geom_1:yshift = -0.6451711006
geom_1:vel_shift = -5.293493449
INFO:DysmalPy:Iter 10 CHI-SQUARE = 3007.851742 DOF = 27860
disk+bulge:total_mass = 10.86834486
disk+bulge:r_eff_disk = 2.028957432
halo:mvirial = 12.82288488
dispprof_LINE:sigma0 = 59.17294407
geom_1:inc = 42.72045022
geom_1:pa = 147.5824829
geom_1:xshift = 0.1035381522
geom_1:yshift = -0.6418476553
geom_1:vel_shift = -5.208883757
INFO:DysmalPy:Iter 11 CHI-SQUARE = 3007.309306 DOF = 27860
disk+bulge:total_mass = 10.84537388
disk+bulge:r_eff_disk = 2.045854719
halo:mvirial = 12.80571025
dispprof_LINE:sigma0 = 59.21611549
geom_1:inc = 44.46906131
geom_1:pa = 147.6156297
geom_1:xshift = 0.1071199159
geom_1:yshift = -0.6446997691
geom_1:vel_shift = -5.280144569
INFO:DysmalPy:Iter 12 CHI-SQUARE = 3007.280107 DOF = 27860
disk+bulge:total_mass = 10.83091615
disk+bulge:r_eff_disk = 2.041894237
halo:mvirial = 12.80019041
dispprof_LINE:sigma0 = 59.07035515
geom_1:inc = 45.44286892
geom_1:pa = 147.5959547
geom_1:xshift = 0.1036601895
geom_1:yshift = -0.6426376258
geom_1:vel_shift = -5.292933151
INFO:DysmalPy:Iter 13 CHI-SQUARE = 3007.236251 DOF = 27860
disk+bulge:total_mass = 10.83865695
disk+bulge:r_eff_disk = 2.036882689
halo:mvirial = 12.80589745
dispprof_LINE:sigma0 = 59.07426415
geom_1:inc = 44.87059754
geom_1:pa = 147.5878073
geom_1:xshift = 0.1032836781
geom_1:yshift = -0.6422652661
geom_1:vel_shift = -5.27681068
INFO:DysmalPy:Iter 14 CHI-SQUARE = 3007.233075 DOF = 27860
disk+bulge:total_mass = 10.84077348
disk+bulge:r_eff_disk = 2.039789375
halo:mvirial = 12.80519019
dispprof_LINE:sigma0 = 59.11641583
geom_1:inc = 44.77529623
geom_1:pa = 147.5990973
geom_1:xshift = 0.1046249005
geom_1:yshift = -0.6431690434
geom_1:vel_shift = -5.281463176
INFO:DysmalPy:Iter 15 CHI-SQUARE = 3007.232618 DOF = 27860
disk+bulge:total_mass = 10.84040688
disk+bulge:r_eff_disk = 2.038689022
halo:mvirial = 12.8051607
dispprof_LINE:sigma0 = 59.10169182
geom_1:inc = 44.78882043
geom_1:pa = 147.5991907
geom_1:xshift = 0.1041919358
geom_1:yshift = -0.6428914038
geom_1:vel_shift = -5.281201733
INFO:DysmalPy:Iter 16 CHI-SQUARE = 3007.232601 DOF = 27860
disk+bulge:total_mass = 10.84031485
disk+bulge:r_eff_disk = 2.038605205
halo:mvirial = 12.80517328
dispprof_LINE:sigma0 = 59.10044981
geom_1:inc = 44.79442182
geom_1:pa = 147.5984447
geom_1:xshift = 0.1041981364
geom_1:yshift = -0.6428959839
geom_1:vel_shift = -5.281632655
INFO:DysmalPy:Iter 17 CHI-SQUARE = 3007.232234 DOF = 27860
disk+bulge:total_mass = 10.84013819
disk+bulge:r_eff_disk = 2.038602824
halo:mvirial = 12.80509821
dispprof_LINE:sigma0 = 59.09937541
geom_1:inc = 44.8062963
geom_1:pa = 147.5982924
geom_1:xshift = 0.104180591
geom_1:yshift = -0.6428869651
geom_1:vel_shift = -5.282021803
INFO:DysmalPy:Iter 18 CHI-SQUARE = 3007.232232 DOF = 27860
disk+bulge:total_mass = 10.84017041
disk+bulge:r_eff_disk = 2.038562767
halo:mvirial = 12.80512703
dispprof_LINE:sigma0 = 59.0991332
geom_1:inc = 44.80369845
geom_1:pa = 147.598199
geom_1:xshift = 0.1041727907
geom_1:yshift = -0.6428811755
geom_1:vel_shift = -5.281928944
INFO:DysmalPy:Iter 19 CHI-SQUARE = 3007.232232 DOF = 27860
disk+bulge:total_mass = 10.84017776
disk+bulge:r_eff_disk = 2.038563995
halo:mvirial = 12.80513077
dispprof_LINE:sigma0 = 59.0992169
geom_1:inc = 44.80322571
geom_1:pa = 147.5982163
geom_1:xshift = 0.1041743573
geom_1:yshift = -0.6428820783
geom_1:vel_shift = -5.281947491
INFO:DysmalPy:
End: 2024-05-02 10:46:38.212010
******************
Time= 24.00 (sec), 0:24.00 (m:s)
MPFIT Status = 1
MPFIT Error/Warning Message = None
******************
plot_model_multid: ndim=3: moment=False
------------------------------------------------------------------
Dysmalpy MPFIT fitting complete for: GS4_43501
output folder: /Users/jespejo/Dropbox/Postdoc/Data/dysmalpy_test_examples/JUPYTER_OUTPUT_3D_FITTING_WRAPPER/MPFIT/
------------------------------------------------------------------
4) Examine results
List all saved files in output directory:
for f in os.listdir(outdir_mpfit): print(f)
GS4_43501_mpfit_bestfit_results_report.info
GS4_43501_LINE_bestfit_velprofile.dat
GS4_43501_mpfit_bestfit_OBS_extract_1D.png
GS4_43501_menc_tot_bary_dm.dat
GS4_43501_mpfit_results.pickle
GS4_43501_OBS_bestfit_cube.fits
GS4_43501_mpfit_bestfit_results.dat
GS4_43501_fit_report.txt
GS4_43501_fitting_3D_mpfit.params
GS4_43501_bestfit_menc.dat
GS4_43501_mpfit_bestfit_OBS_spaxels.png
GS4_43501_mpfit.log
GS4_43501_mpfit_bestfit_OBS_apertures.png
GS4_43501_OBS_out-cube.fits
GS4_43501_model.pickle
GS4_43501_vcirc_tot_bary_dm.dat
GS4_43501_mpfit_bestfit_OBS_extract_2D.png
GS4_43501_mpfit_bestfit_OBS_channels.png
GS4_43501_bestfit_vcirc.dat
Result plots
Read in parameter file
params = utils_io.read_fitting_params(fname=param_filename)
# Override data + output paths:
params['datadir'] = datadir
params['outdir'] = outdir_mpfit
# Add the plot type:
params['plot_type'] = plot_type
f_galmodel = params['outdir'] + '{}_model.pickle'.format(params['galID'])
f_results = params['outdir'] + '{}_{}_results.pickle'.format(params['galID'],
params['fit_method'])
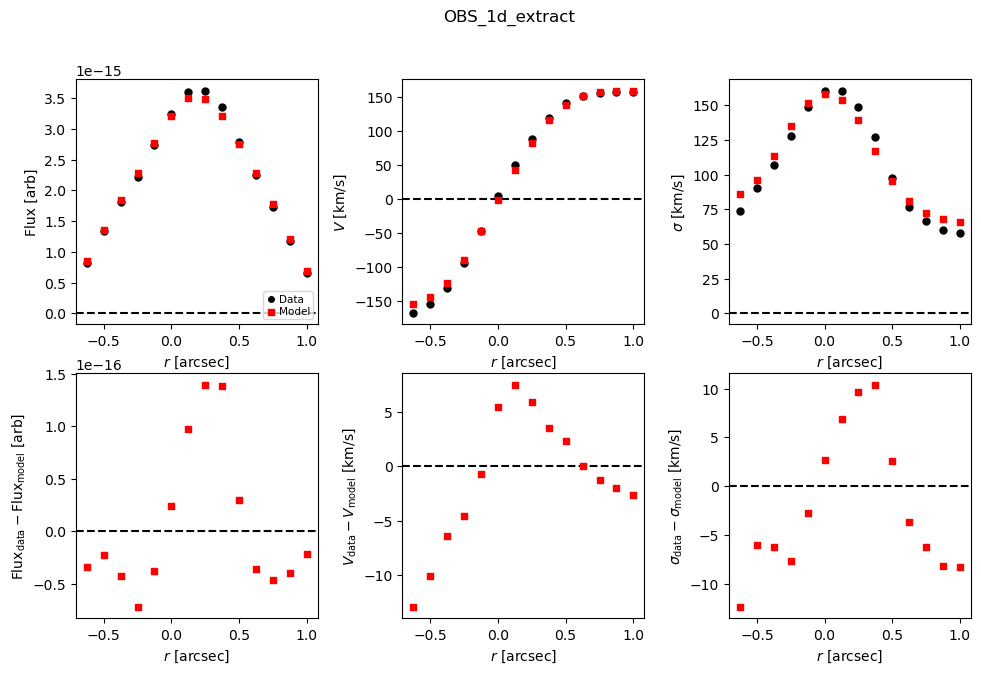
Best-fit plot (extracted to 1D circular apertures and 2D maps):
# Look at best-fit:
filepath = outdir_mpfit+"{}_{}_bestfit_{}_extract_1D.{}".format(params['galID'],
params['fit_method'],
params['obs_1_name'],
params['plot_type'])
Image(filename=filepath, width=600)

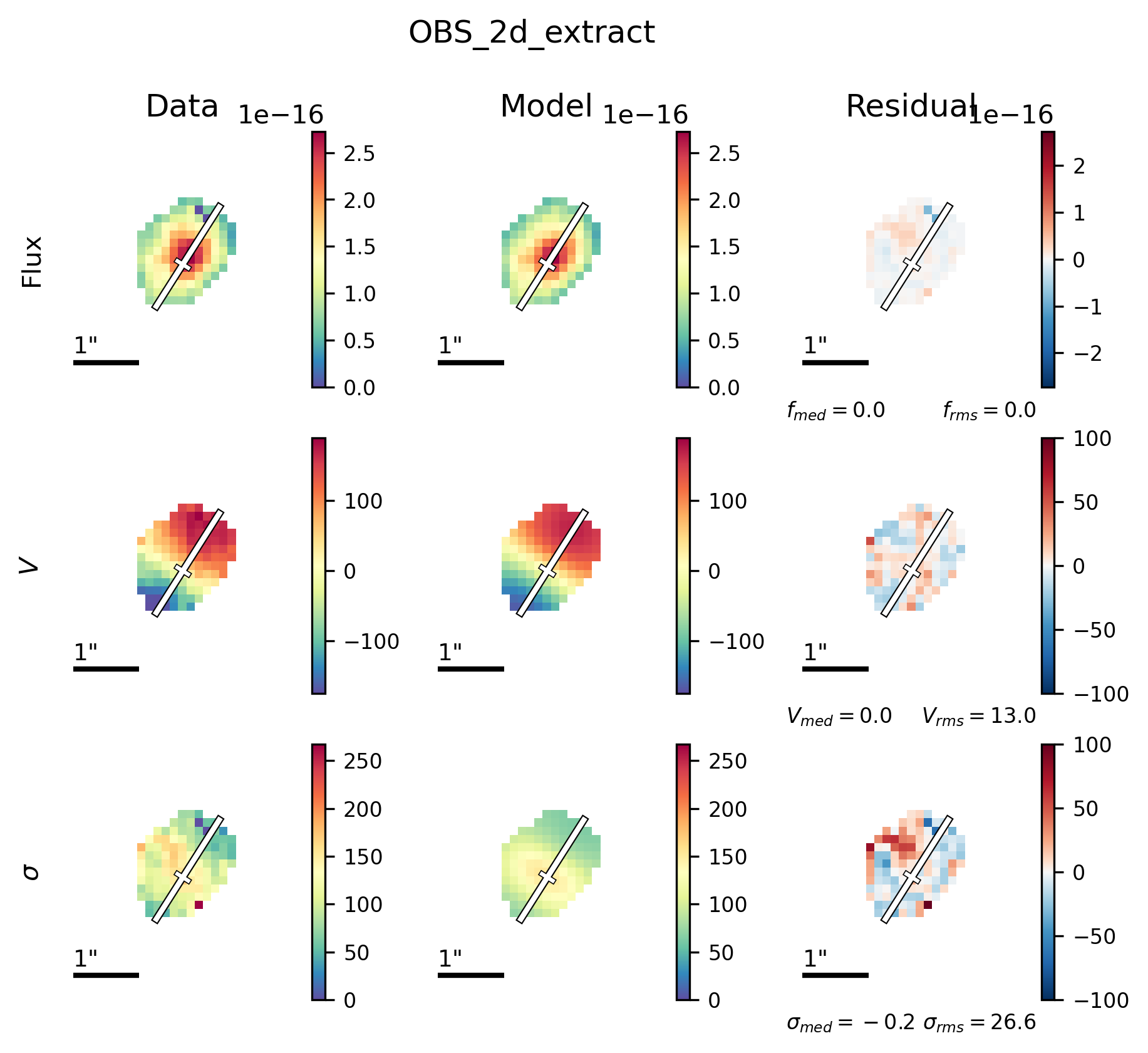
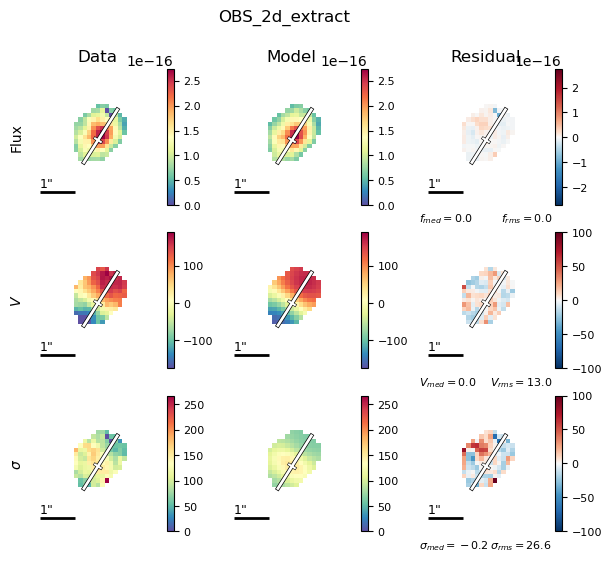
filepath = outdir_mpfit+"{}_{}_bestfit_{}_extract_2D.{}".format(params['galID'],
params['fit_method'],
params['obs_1_name'],
params['plot_type'])
Image(filename=filepath, width=600)
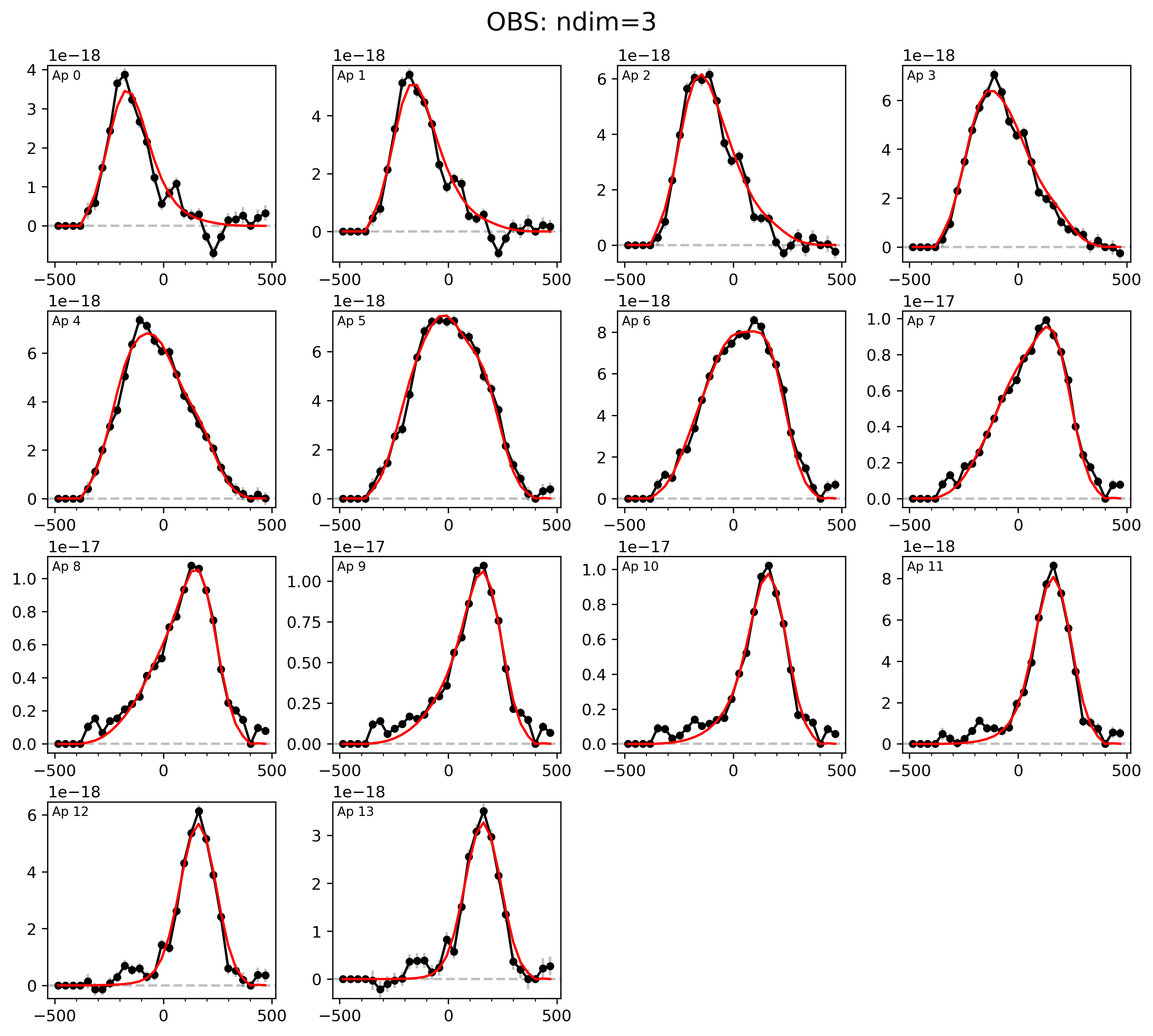
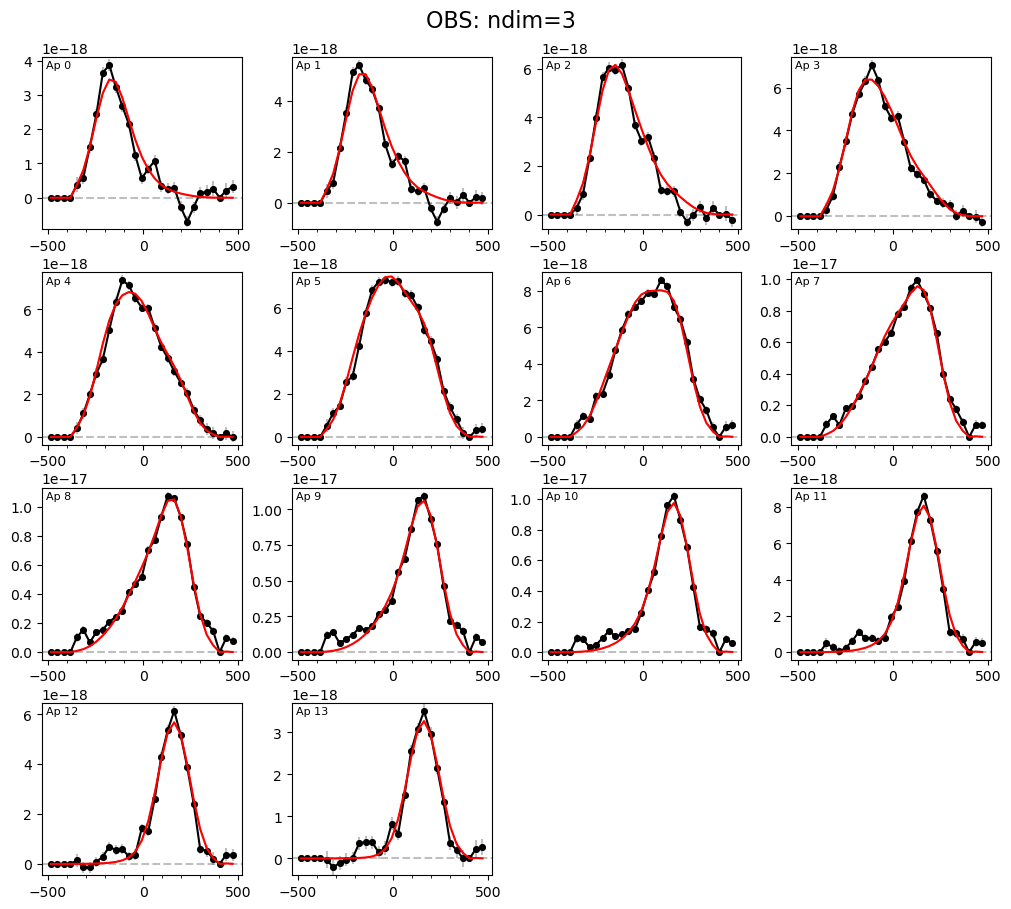
LOS velocity distribution in each of the 1D circular apertures:
# Look at per-aperture profiles:
filepath = outdir_mpfit+"{}_{}_bestfit_{}_apertures.{}".format(params['galID'],
params['fit_method'],
params['obs_1_name'],
params['plot_type'])
Image(filename=filepath, width=600)
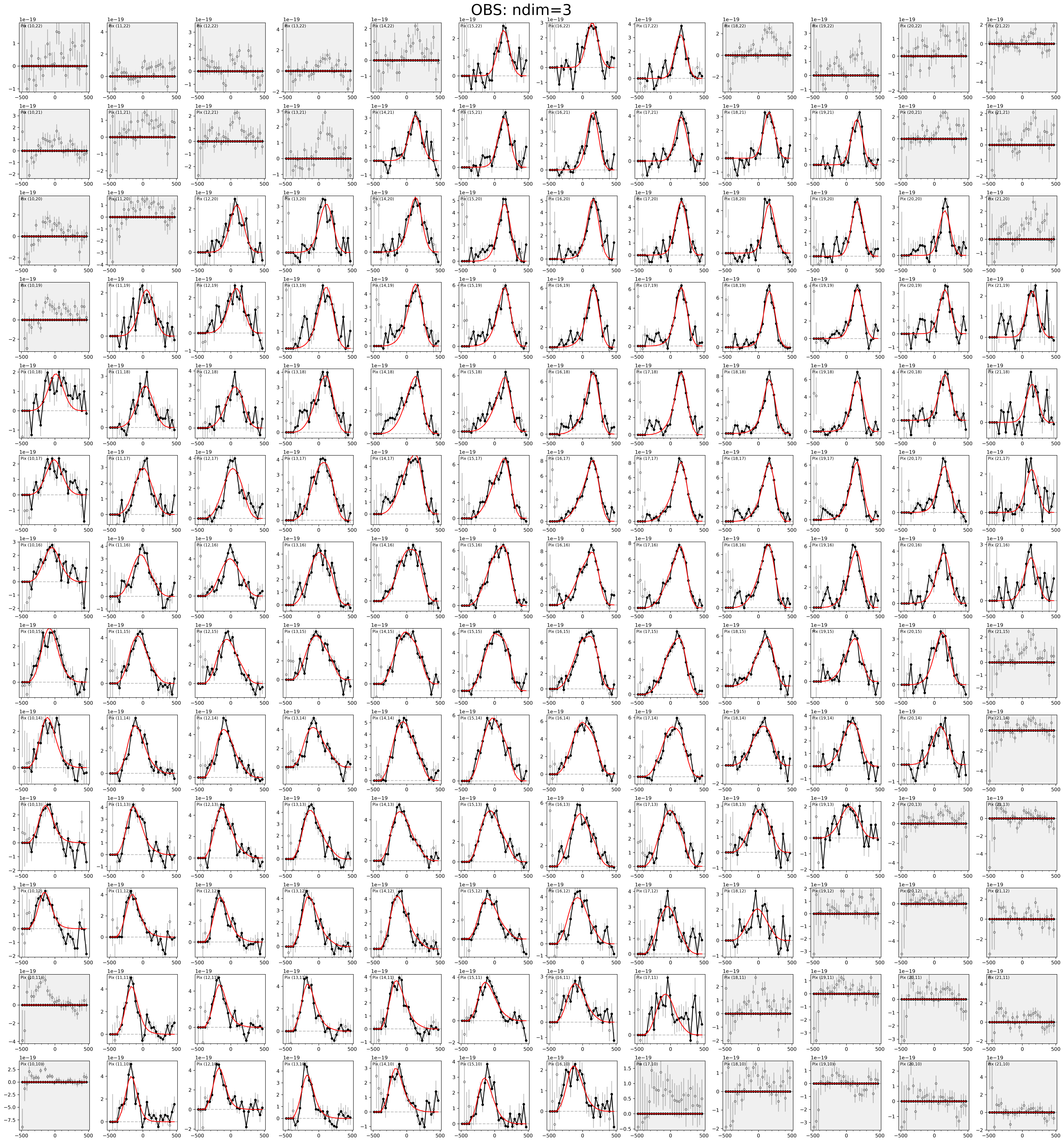
LOS velocity distribution in each spaxel:
# Look at per-spaxel profiles:
filepath = outdir_mpfit+"{}_{}_bestfit_{}_spaxels.{}".format(params['galID'],
params['fit_method'],
params['obs_1_name'],
params['plot_type'])
Image(filename=filepath, width=600)
Channel maps:
# Look at channel maps:
filepath = outdir_mpfit+"{}_{}_bestfit_{}_channels.{}".format(params['galID'],
params['fit_method'],
params['obs_1_name'],
params['plot_type'])
Image(filename=filepath, width=600)

Directly generating result plots
Reload the galaxy, results files:
gal, results = fitting.reload_all_fitting(filename_galmodel=f_galmodel,
filename_results=f_results,
fit_method=params['fit_method'])
Plot the best-fit results:
results.plot_results(gal)
plot_model_multid: ndim=3: moment=False

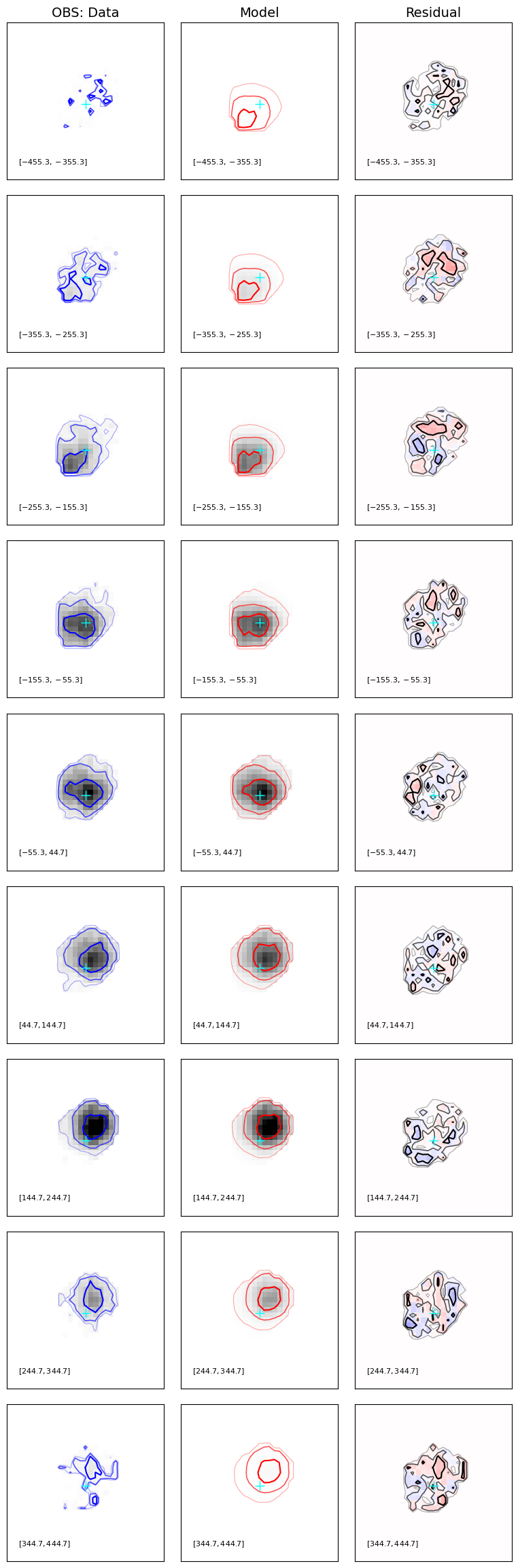
Results reports
We now look at the results reports, which include the best-fit values and uncertainties (as well as other fitting settings and output).
# Print report
print(results.results_report(gal=gal))
###############################
Fitting for GS4_43501
Date: 2024-05-02 10:47:05.749237
obs: OBS
Datafiles:
zcalc_truncate: True
n_wholepix_z_min: 3
oversample: 1
oversize: 1
Fitting method: MPFIT
fit status: 1
pressure_support: True
pressure_support_type: 1
###############################
Fitting results
-----------
disk+bulge
total_mass 10.8402 +/- 0.0915
r_eff_disk 2.0386 +/- 0.0685
n_disk 1.0000 [FIXED]
r_eff_bulge 1.0000 [FIXED]
n_bulge 4.0000 [FIXED]
bt 0.3000 [FIXED]
mass_to_light 1.0000 [FIXED]
noord_flat True
-----------
halo
mvirial 12.8051 +/- 0.1290
fdm 0.1593 [TIED]
conc 5.0000 [FIXED]
-----------
dispprof_LINE
sigma0 59.0993 +/- 1.5962
-----------
zheightgaus
sigmaz 0.3463 [TIED]
-----------
geom_1
inc 44.8022 +/- 6.3949
pa 147.5982 +/- 0.6322
xshift 0.1042 +/- 0.0348
yshift -0.6429 +/- 0.0252
vel_shift -5.2819 +/- 1.0155
-----------
Adiabatic contraction: False
-----------
Red. chisq: 1.0563
To directly save the results report to a file, we can use the following:
# Save report to file:
f_report = params['outdir'] + '{}_fit_report.txt'.format(params['galID'])
results.results_report(gal=gal, filename=f_report)
Also note the fitting wrappers automatically save two versions of the report files:
fbase = '{}_{}_bestfit_results'.format(params['galID'], params['fit_method'])
f_report_pretty = params['outdir'] + fbase + '_report.info'
f_report_machine = params['outdir'] + fbase + '.dat'
The “pretty” version, automatically saved as *_best_fit_results_report.info, is formatted to be human-readable, and includes more information on the fit settings at the beginning (for reference).
with open(f_report_pretty, 'r') as f:
lines = [line.rstrip() for line in f]
for line in lines: print(line)
###############################
Fitting for GS4_43501
Date: 2024-05-02 10:46:54.021042
obs: OBS
Datafiles:
zcalc_truncate: True
n_wholepix_z_min: 3
oversample: 1
oversize: 1
Fitting method: MPFIT
fit status: 1
pressure_support: True
pressure_support_type: 1
###############################
Fitting results
-----------
disk+bulge
total_mass 10.8402 +/- 0.0915
r_eff_disk 2.0386 +/- 0.0685
n_disk 1.0000 [FIXED]
r_eff_bulge 1.0000 [FIXED]
n_bulge 4.0000 [FIXED]
bt 0.3000 [FIXED]
mass_to_light 1.0000 [FIXED]
noord_flat True
-----------
halo
mvirial 12.8051 +/- 0.1290
fdm 0.1593 [TIED]
conc 5.0000 [FIXED]
-----------
dispprof_LINE
sigma0 59.0993 +/- 1.5962
-----------
zheightgaus
sigmaz 0.3463 [TIED]
-----------
geom_1
inc 44.8022 +/- 6.3949
pa 147.5982 +/- 0.6322
xshift 0.1042 +/- 0.0348
yshift -0.6429 +/- 0.0252
vel_shift -5.2819 +/- 1.0155
-----------
Adiabatic contraction: False
-----------
Red. chisq: 1.0563
The “machine” version, automatically saved as *_best_fit_results.dat, is formatted as a machine-readable space-separated ascii file. It includes key parameter fit information, as well as the best-fit reduced chisq.
with open(f_report_machine, 'r') as f:
lines = [line.rstrip() for line in f]
for line in lines: print(line)
# component param_name fixed best_value l68_err u68_err
disk+bulge total_mass False 10.8402 0.0915 0.0915
disk+bulge r_eff_disk False 2.0386 0.0685 0.0685
disk+bulge n_disk True 1.0000 -99.0000 -99.0000
disk+bulge r_eff_bulge True 1.0000 -99.0000 -99.0000
disk+bulge n_bulge True 4.0000 -99.0000 -99.0000
disk+bulge bt True 0.3000 -99.0000 -99.0000
disk+bulge mass_to_light True 1.0000 -99.0000 -99.0000
halo mvirial False 12.8051 0.1290 0.1290
halo fdm TIED 0.1593 -99.0000 -99.0000
halo conc True 5.0000 -99.0000 -99.0000
dispprof_LINE sigma0 False 59.0993 1.5962 1.5962
zheightgaus sigmaz TIED 0.3463 -99.0000 -99.0000
geom_1 inc False 44.8022 6.3949 6.3949
geom_1 pa False 147.5982 0.6322 0.6322
geom_1 xshift False 0.1042 0.0348 0.0348
geom_1 yshift False -0.6429 0.0252 0.0252
geom_1 vel_shift False -5.2819 1.0155 1.0155
mvirial ----- ----- 12.8051 -99.0000 -99.0000
fit_status ----- ----- 1 -99.0000 -99.0000
adiab_contr ----- ----- False -99.0000 -99.0000
redchisq ----- ----- 1.0563 -99.0000 -99.0000
noord_flat ----- ----- True -99.0000 -99.0000
pressure_support ----- ----- True -99.0000 -99.0000
pressure_support_type ----- ----- 1 -99.0000 -99.0000